Custom mean functions: metalearning with GPs#
One of the advantages of Gaussian process is their flexibility as a modeling tool. For instance, if the modeler knows that there is an underlying trend in the data, they can specify a mean function that captures this trend.
In this notebook, we illustrate how to use GPflow to construct a custom neural network mean function for GPs that can capture complex trends. We look at this functionality in the context of metalearning, where a number of metatasks are available at train time and the user wants to adapt a flexible model to new tasks at test time.
For an in-depth discussion on this topic, see (Fortuin and Rätsch, 2019). This notebook reproduces section 4.2 of this paper.
[1]:
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import gpflow
from gpflow.ci_utils import reduce_in_tests
from gpflow.kernels import RBF
from gpflow.models import GPR
# for reproducibility of this notebook:
np.random.seed(1)
tf.random.set_seed(2)
%matplotlib inline
2023-08-09 12:27:18.060076: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Could not find TensorRT
Generate the tasks#
To generate the meta and test tasks, we sample from a Gaussian process with an Squared Exponential covariance function and a sinusoidal mean function. Each task is a realization of this process.
[2]:
def generate_data(num_functions=10, N=500):
"""
For each function, sample the value at `N` equally spaced
points in the [−5, 5] interval (Fortuin and Rätsch, 2019).
Returns:
Tuple of np.arrays of size (N, 1) and (N, num_functions).
"""
jitter = 1e-6
Xs = np.linspace(-5.0, 5.0, N)[:, None]
kernel = RBF(lengthscales=1.0)
cov = kernel(Xs)
L = np.linalg.cholesky(cov + np.eye(N) * jitter)
epsilon = np.random.randn(N, num_functions)
F = np.sin(Xs) + np.matmul(L, epsilon)
return Xs, F
We generate 10 tasks for illustration.
[3]:
Xs, F = generate_data(10)
[4]:
plt.plot(Xs, F)
We generate the meta and test tasks.
[5]:
def generate_meta_and_test_tasks(num_datapoints, num_meta, num_test, N):
"""Generates meta-task datasets {D_i} = {x_i, y_i} and target task training
and test data {\tilde{x}, \tilde{y}} (Fortuin and Rätsch, 2019).
Args:
num_datapoints: The number of training points, \tilde{n} in Table S1.
num_meta: The number of meta-tasks ("sampled functions").
num_test: The number of test tasks ("unseen functions").
N: Number of sampled data points per function.
Returns:
A tuple (meta, test) where
- meta: List of num_meta pairs of arrays (X, Y) of size (n, 1) each.
- test: List of num_test pairs of pairs of arrays of sizes
(((num_datapoints, 1), (num_datapoints, 1)),
((N - num_datapoints, 1), (N - num_datapoints, 1))).
"""
Xs, F = generate_data(num_functions=num_meta + num_test, N=N)
meta = []
sd = 1e-1 # Standard deviation for normally-distributed observation noise.
for i in range(num_meta):
# We always use all data points of the curve to train the mean function,
# i.e. n_i = N.
noise = sd * np.random.randn(N, 1)
Y = F[:, i][:, None] + noise
meta.append((Xs, Y))
test = []
rand_indices = np.random.permutation(N)
train_i = sorted(rand_indices[:num_datapoints])
test_i = sorted(rand_indices[num_datapoints:])
for i in range(num_test):
# Form target training set, \tilde{D}_{train} (see Figure 1).
noise = sd * np.random.randn(num_datapoints, 1)
train_X = Xs[train_i]
train_Y = F[train_i, num_meta + i][:, None] + noise
# Form target test set, \tilde{D}_{test}, which is disjoint from the
# target traininig set as it is in the original implementation.
test_X = Xs[test_i]
test_Y = F[test_i, num_meta + i][:, None]
# Form target tasks' datasets \tilde{D} as a pair of pairs,
# (\tilde{X}, \tilde{Y}) and (\tilde{X}*, \tilde{Y}*).
test.append(((train_X, train_Y), (test_X, test_Y)))
return meta, test
[6]:
num_datapoints = 5
# Although the original experiment uses (1000, 200, 50) for the following
# parameters, we will use (50, 10, 500) for computational reasons.
num_meta_tasks = reduce_in_tests(50)
num_test_tasks = reduce_in_tests(10)
num_data_per_task = 500
num_iter = reduce_in_tests(5)
meta, test = generate_meta_and_test_tasks(
num_datapoints, num_meta_tasks, num_test_tasks, num_data_per_task
)
Create the mean function#
We will use a Keras model Deep Neural Network as mean function.
[7]:
from gpflow.config import default_float
def build_mean_function():
tf.keras.backend.set_floatx("float64")
assert default_float() == np.float64
inputs = tf.keras.layers.Input(shape=(1,))
x = tf.keras.layers.Dense(64, activation="relu")(inputs)
x = tf.keras.layers.Dense(64, activation="relu")(x)
outputs = tf.keras.layers.Dense(1)(x)
return tf.keras.Model(inputs=inputs, outputs=outputs)
Build the GP metamodel#
Metalearning boils down to learning a good prior that can generalize to new tasks with a small number of data points. This framework is prevalent in GP modeling, where we usually maximize the marginal likelihood to learn a good set of hyperparameters that specify the GP prior.
We perform the same optimization here, while sharing the hyperparameters across all the metatasks. For simplicity, we fix the kernel and likelihood parameters and learn those only for the mean function. Hence, our metalearning procedure is to cycle through the metatasks continuously, optimizing their marginal likelihood until a convergence criteria is reached (here, we just implement a fixed number of iterations over the tasks).
To begin this process, first we create a utility function that takes in a task (X, Y) and a mean function and outputs a GP model.
[8]:
from gpflow import set_trainable
def build_model(data, mean_function):
model = GPR(data, kernel=RBF(), mean_function=mean_function)
set_trainable(model.kernel, False)
model.likelihood.variance.assign(1e-2)
set_trainable(model.likelihood, False)
return model
[9]:
def create_optimization_step(optimizer, model: gpflow.models.GPR):
@tf.function
def optimization_step():
with tf.GradientTape(watch_accessed_variables=False) as tape:
tape.watch(model.trainable_variables)
objective = model.training_loss()
grads = tape.gradient(objective, model.trainable_variables)
optimizer.apply_gradients(zip(grads, model.trainable_variables))
return objective
return optimization_step
def run_adam(model, iterations):
"""
Utility function running the Adam optimizer
:param model: GPflow model
:param interations: number of iterations
"""
# Create an Adam Optimizer action
logf = []
adam = tf.optimizers.Adam()
optimization_step = create_optimization_step(adam, model)
for step in range(iterations):
loss = optimization_step()
if step % 10 == 0:
# print("num variables", len(model.trainable_variables))
logf.append(loss.numpy())
return logf
Next, we define the training loop for metalearning.
[10]:
import time
def train_loop(meta_tasks):
"""
Metalearning training loop. Trained for 100 epochs in original experiment.
:param meta_tasks: list of metatasks.
:param num_iter: number of iterations of tasks set
:returns: a mean function object
"""
# Initialize mean function
mean_function = build_mean_function()
# Iterate for several passes over the tasks set
for iteration in range(num_iter):
ts = time.time()
print("Currently in meta-iteration/epoch {}".format(iteration))
# Iterate over tasks
for i, task in enumerate(meta_tasks):
data = task # (X, Y)
model = build_model(data, mean_function=mean_function)
run_adam(model, reduce_in_tests(100))
print(">>>> iteration took {:.2f} s".format(time.time() - ts))
return mean_function
mean_function_optimal = train_loop(meta)
Currently in meta-iteration/epoch 0
>>>> iteration took 71.46 s
Currently in meta-iteration/epoch 1
>>>> iteration took 73.52 s
Currently in meta-iteration/epoch 2
>>>> iteration took 73.37 s
Currently in meta-iteration/epoch 3
>>>> iteration took 71.55 s
Currently in meta-iteration/epoch 4
>>>> iteration took 73.04 s
Finally, we use the optimized mean function for all of the test tasks. NOTE: We do not do any further optimization for the hyperparameters in this step.
[11]:
test_models = [build_model(data, mean_function_optimal) for (data, _) in test]
Assess the model#
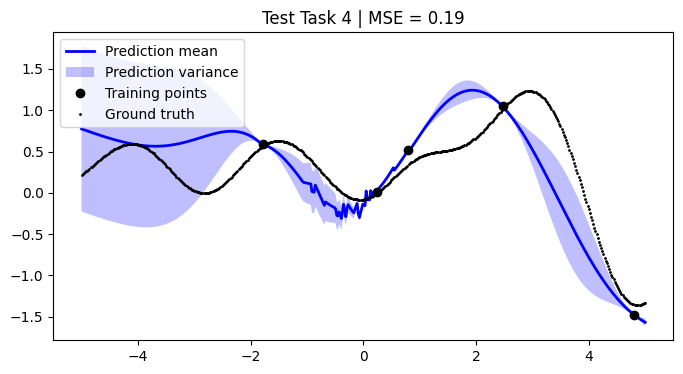
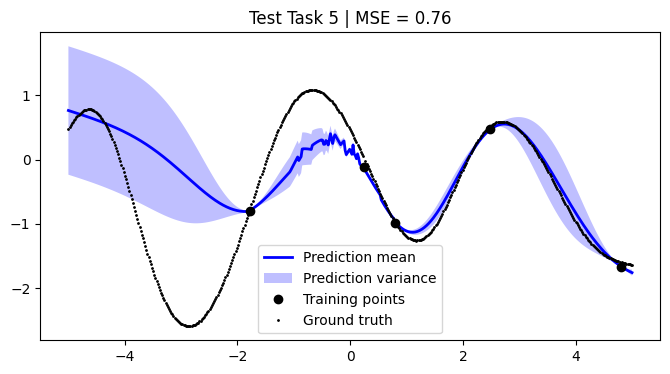
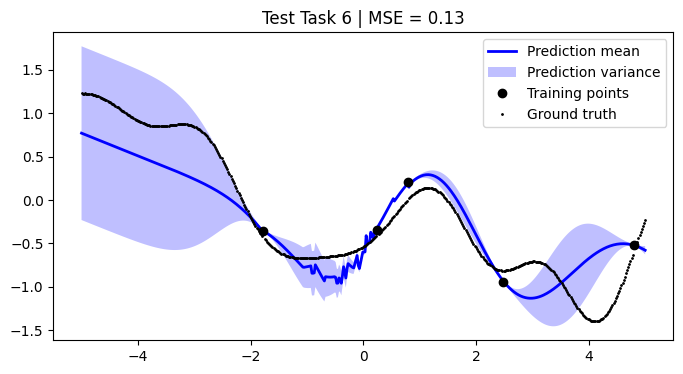
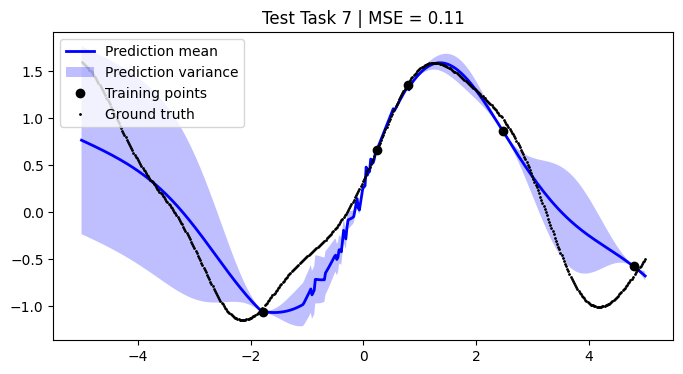
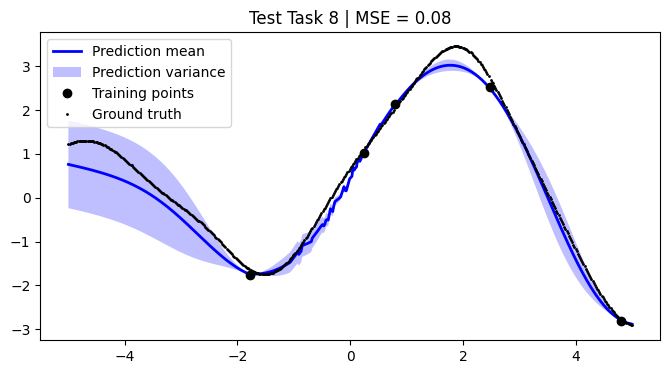
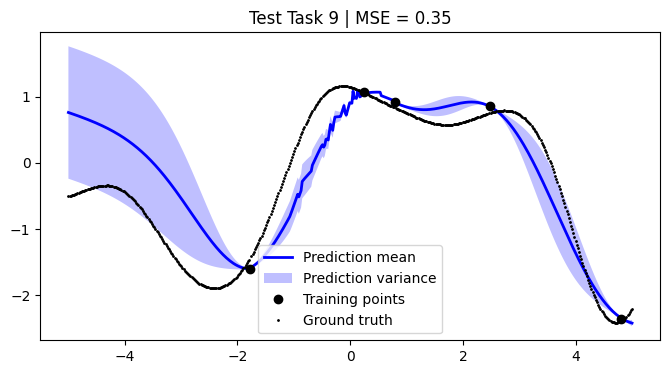
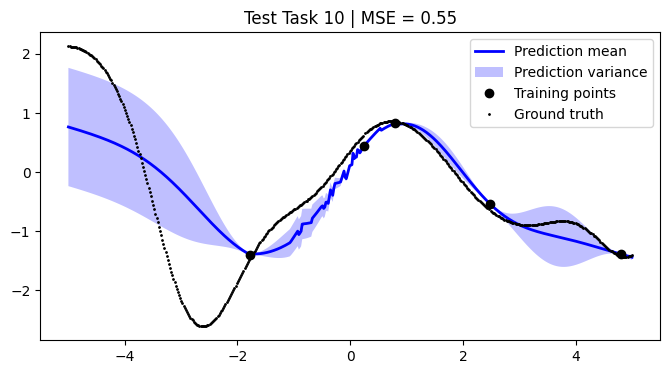
We assess the performance of this procedure on the test tasks. For this, we use the mean squared error as a performance metric.
[12]:
def mean_squared_error(y, y_pred):
return np.mean((y - y_pred) ** 2)
[13]:
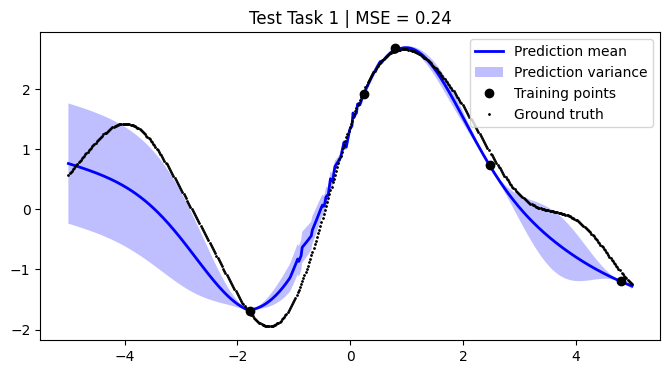
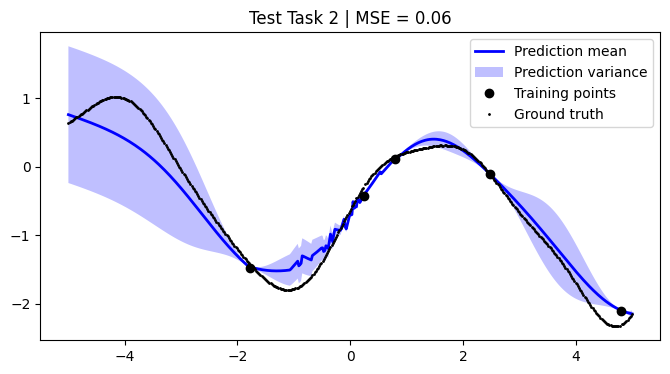
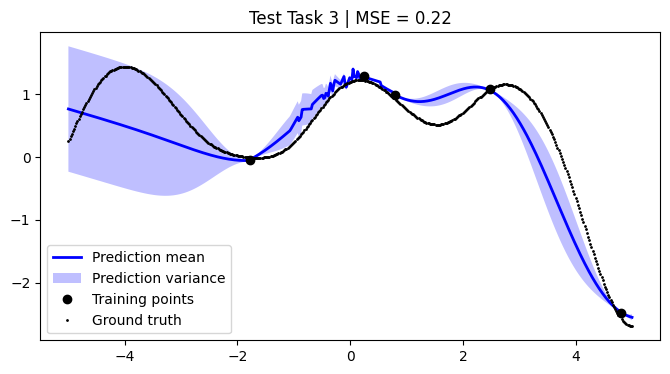
mean_squared_errors = []
for i, test_task in enumerate(test):
plt.figure(figsize=(8, 4))
(train_X, train_Y), (Xs, F) = test_task
pred_mean, pred_var = test_models[i].predict_f(Xs)
plt.plot(Xs, pred_mean, label="Prediction mean", color="blue", linewidth=2)
plt.fill_between(
Xs.squeeze(1),
tf.squeeze(pred_mean - pred_var),
tf.squeeze(pred_mean + pred_var),
alpha=0.25,
facecolor="blue",
label="Prediction variance",
)
plt.plot(train_X, train_Y, "ko", label="Training points")
plt.plot(Xs, F, "ko", label="Ground truth", linewidth=2, markersize=1)
mse = mean_squared_error(F, pred_mean)
mean_squared_errors.append(mse)
plt.title(f"Test Task {i + 1} | MSE = {mse:.2f}")
plt.legend()
plt.show()
[14]:
mean_mse = np.mean(mean_squared_errors)
std_mse = np.std(mean_squared_errors) / np.sqrt(num_test_tasks)
print(
f"The mean MSE over all {num_test_tasks} test tasks is {mean_mse:.2f} +/- {std_mse:.2f}"
)
The mean MSE over all 10 test tasks is 0.27 +/- 0.07
We achieve comparable results to those reported in the paper.
NOTE: We use only 50 metatasks and 10 test tasks over 5 epochs for scalability, whereas the paper uses 1,000 and 200 respectively over 100 epochs. To compensate, we sample 500 points per curve, whereas the paper samples only 50 points. Hence, there might be some discrepancies in the results.
References#
Fortuin, Vincent, and Gunnar Rätsch. “Deep Mean Functions for Meta-Learning in Gaussian Processes.” arXiv preprint arXiv:1901.08098 (2019).