Heteroskedastic Likelihood and Multi-Latent GP#
Standard (Homoskedastic) Regression#
In standard GP regression, the GP latent function is used to learn the location parameter of a likelihood distribution (usually a Gaussian) as a function of the input \(x\), whereas the scale parameter is considered constant. This is a homoskedastic model, which is unable to capture variations of the noise distribution with the input \(x\).
Heteroskedastic Regression#
This notebooks shows how to construct a model which uses multiple (here two) GP latent functions to learn both the location and the scale of the Gaussian likelihood distribution. It does so by connecting a Multi-Output Kernel, which generates multiple GP latent functions, to a Heteroskedastic Likelihood, which maps the latent GPs into a single likelihood.
The generative model is described as:
The function \(\text{transform}\) is used to map from the unconstrained GP \(f_2\) to positive-only values, which is required as it represents the \(\text{scale}\) of a Gaussian likelihood. In this notebook, the \(\exp\) function will be used as the \(\text{transform}\). Other positive transforms such as the \(\text{softplus}\) function can also be used.
[1]:
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import tensorflow_probability as tfp
import gpflow as gpf
2022-05-10 10:56:00.523700: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-05-10 10:56:00.523730: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
Data Generation#
We generate heteroskedastic data by substituting the random latent functions \(f_1\) and \(f_2\) of the generative model by deterministic \(\sin\) and \(\cos\) functions. The input \(X\) is built with \(N=1001\) uniformly spaced values in the interval \([0, 4\pi]\). The outputs \(Y\) are still sampled from a Gaussian likelihood.
[2]:
N = 1001
np.random.seed(0)
tf.random.set_seed(0)
# Build inputs X
X = np.linspace(0, 4 * np.pi, N)[:, None] # X must be of shape [N, 1]
# Deterministic functions in place of latent ones
f1 = np.sin
f2 = np.cos
# Use transform = exp to ensure positive-only scale values
transform = np.exp
# Compute loc and scale as functions of input X
loc = f1(X)
scale = transform(f2(X))
# Sample outputs Y from Gaussian Likelihood
Y = np.random.normal(loc, scale)
Plot Data#
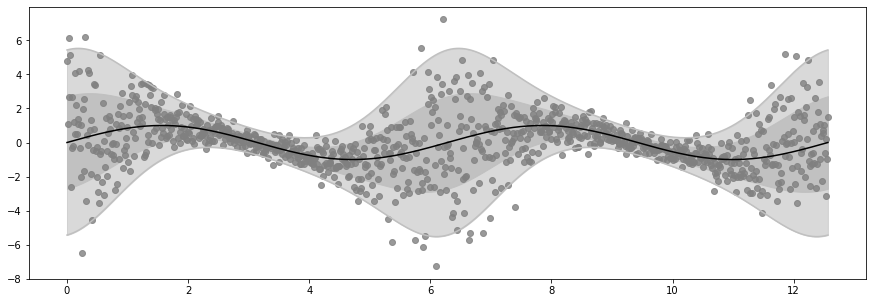
Note how the distribution density (shaded area) and the outputs \(Y\) both change depending on the input \(X\).
[3]:
def plot_distribution(X, Y, loc, scale):
plt.figure(figsize=(15, 5))
x = X.squeeze()
for k in (1, 2):
lb = (loc - k * scale).squeeze()
ub = (loc + k * scale).squeeze()
plt.fill_between(x, lb, ub, color="silver", alpha=1 - 0.05 * k ** 3)
plt.plot(x, lb, color="silver")
plt.plot(x, ub, color="silver")
plt.plot(X, loc, color="black")
plt.scatter(X, Y, color="gray", alpha=0.8)
plt.show()
plt.close()
plot_distribution(X, Y, loc, scale)
Build Model#
Likelihood#
This implements the following part of the generative model:
[4]:
likelihood = gpf.likelihoods.HeteroskedasticTFPConditional(
distribution_class=tfp.distributions.Normal, # Gaussian Likelihood
scale_transform=tfp.bijectors.Exp(), # Exponential Transform
)
print(f"Likelihood's expected latent_dim: {likelihood.latent_dim}")
Likelihood's expected latent_dim: 2
2022-05-10 10:56:02.799816: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-05-10 10:56:02.799850: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2022-05-10 10:56:02.799870: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (49c966262641): /proc/driver/nvidia/version does not exist
2022-05-10 10:56:02.800307: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
Kernel#
This implements the following part of the generative model:
with both kernels being modeled as separate and independent \(\text{SquaredExponential}\) kernels.
[5]:
kernel = gpf.kernels.SeparateIndependent(
[
gpf.kernels.SquaredExponential(), # This is k1, the kernel of f1
gpf.kernels.SquaredExponential(), # this is k2, the kernel of f2
]
)
# The number of kernels contained in gpf.kernels.SeparateIndependent must be the same as likelihood.latent_dim
Inducing Points#
Since we will use the SVGP model to perform inference, we need to implement the inducing variables \(U_1\) and \(U_2\), both with size \(M=20\), which are used to approximate \(f_1\) and \(f_2\) respectively, and initialize the inducing points positions \(Z_1\) and \(Z_2\). This gives a total of \(2M=40\) inducing variables and inducing points.
The inducing variables and their corresponding inputs will be Separate and Independent, but both \(Z_1\) and \(Z_2\) will be initialized as \(Z\), which are placed as \(M=20\) equally spaced points in \([\min(X), \max(X)]\).
[6]:
M = 20 # Number of inducing variables for each f_i
# Initial inducing points position Z
Z = np.linspace(X.min(), X.max(), M)[:, None] # Z must be of shape [M, 1]
inducing_variable = gpf.inducing_variables.SeparateIndependentInducingVariables(
[
gpf.inducing_variables.InducingPoints(Z), # This is U1 = f1(Z1)
gpf.inducing_variables.InducingPoints(Z), # This is U2 = f2(Z2)
]
)
SVGP Model#
Build the SVGP model by composing the Kernel, the Likelihood and the Inducing Variables.
Note that the model needs to be instructed about the number of latent GPs by passing num_latent_gps=likelihood.latent_dim.
[7]:
model = gpf.models.SVGP(
kernel=kernel,
likelihood=likelihood,
inducing_variable=inducing_variable,
num_latent_gps=likelihood.latent_dim,
)
model
[7]:
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| SVGP.kernel.kernels[0].variance | Parameter | Softplus | True | () | float64 | 1.0 | |
| SVGP.kernel.kernels[0].lengthscales | Parameter | Softplus | True | () | float64 | 1.0 | |
| SVGP.kernel.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1.0 | |
| SVGP.kernel.kernels[1].lengthscales | Parameter | Softplus | True | () | float64 | 1.0 | |
| SVGP.inducing_variable.inducing_variable_list[0].Z | Parameter | Identity | True | (20, 1) | float64 | [[0.... | |
| SVGP.inducing_variable.inducing_variable_list[1].Z | Parameter | Identity | True | (20, 1) | float64 | [[0.... | |
| SVGP.q_mu | Parameter | Identity | True | (20, 2) | float64 | [[0., 0.... | |
| SVGP.q_sqrt | Parameter | FillTriangular | True | (2, 20, 20) | float64 | [[[1., 0., 0.... |
Model Optimization#
Build Optimizers (NatGrad + Adam)#
[8]:
data = (X, Y)
loss_fn = model.training_loss_closure(data)
gpf.utilities.set_trainable(model.q_mu, False)
gpf.utilities.set_trainable(model.q_sqrt, False)
variational_vars = [(model.q_mu, model.q_sqrt)]
natgrad_opt = gpf.optimizers.NaturalGradient(gamma=0.1)
adam_vars = model.trainable_variables
adam_opt = tf.optimizers.Adam(0.01)
@tf.function
def optimisation_step():
natgrad_opt.minimize(loss_fn, variational_vars)
adam_opt.minimize(loss_fn, adam_vars)
2022-05-10 10:56:02.893655: W tensorflow/python/util/util.cc:368] Sets are not currently considered sequences, but this may change in the future, so consider avoiding using them.
Run Optimization Loop#
[9]:
epochs = 100
log_freq = 20
for epoch in range(1, epochs + 1):
optimisation_step()
# For every 'log_freq' epochs, print the epoch and plot the predictions against the data
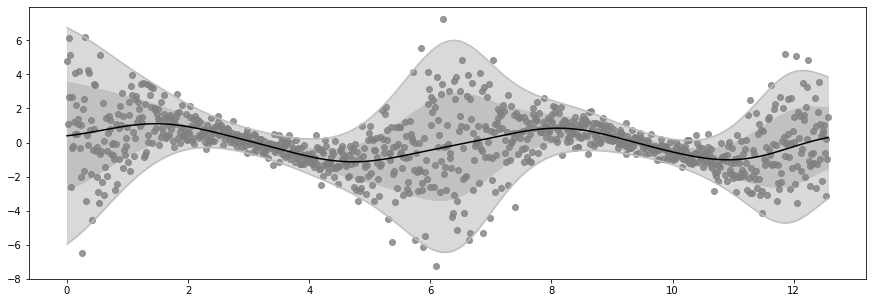
if epoch % log_freq == 0 and epoch > 0:
print(f"Epoch {epoch} - Loss: {loss_fn().numpy() : .4f}")
Ymean, Yvar = model.predict_y(X)
Ymean = Ymean.numpy().squeeze()
Ystd = tf.sqrt(Yvar).numpy().squeeze()
plot_distribution(X, Y, Ymean, Ystd)
model
WARNING:tensorflow:AutoGraph could not transform <function separate_independent_conditional_implementation at 0x7f26c6e20040> and will run it as-is.
Please report this to the TensorFlow team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output.
Cause: annotated name 'base_conditional_args_to_map' can't be nonlocal (__autograph_generated_filesjwtqzfo.py, line 39)
To silence this warning, decorate the function with @tf.autograph.experimental.do_not_convert
WARNING: AutoGraph could not transform <function separate_independent_conditional_implementation at 0x7f26c6e20040> and will run it as-is.
Please report this to the TensorFlow team. When filing the bug, set the verbosity to 10 (on Linux, `export AUTOGRAPH_VERBOSITY=10`) and attach the full output.
Cause: annotated name 'base_conditional_args_to_map' can't be nonlocal (__autograph_generated_filesjwtqzfo.py, line 39)
To silence this warning, decorate the function with @tf.autograph.experimental.do_not_convert
WARNING:tensorflow:From /home/circleci/project/.venv/lib/python3.10/site-packages/tensorflow/python/util/deprecation.py:616: calling map_fn_v2 (from tensorflow.python.ops.map_fn) with dtype is deprecated and will be removed in a future version.
Instructions for updating:
Use fn_output_signature instead
Epoch 20 - Loss: 1471.8678
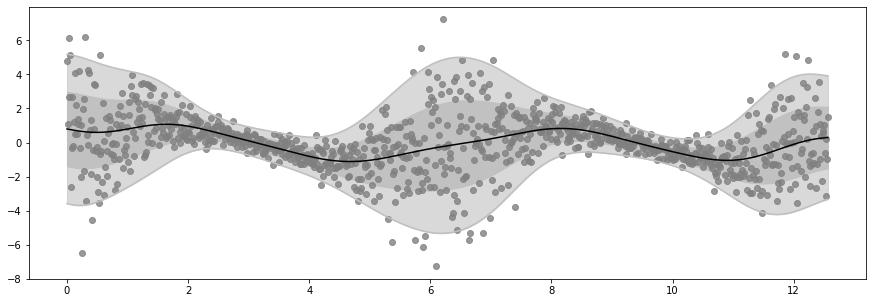
Epoch 40 - Loss: 1452.4426
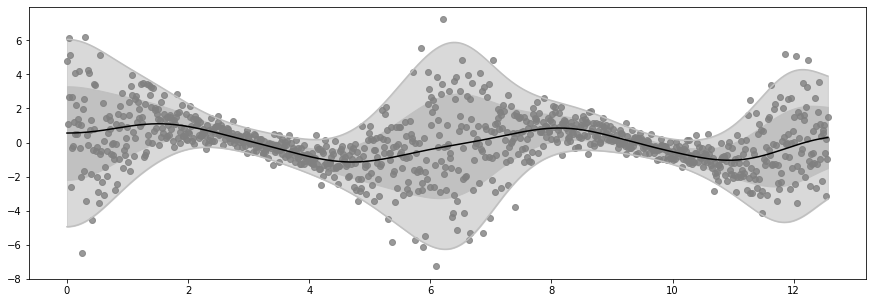
Epoch 60 - Loss: 1450.7381
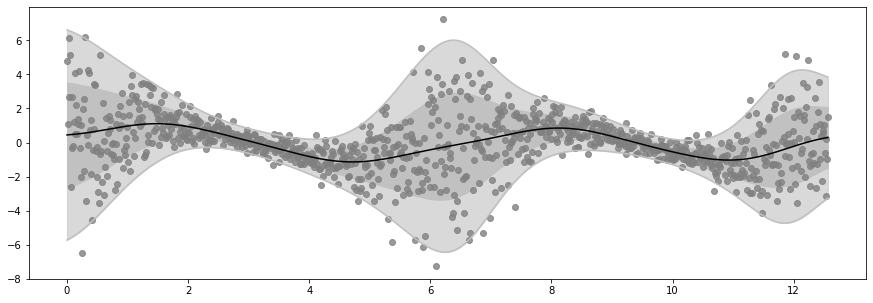
Epoch 80 - Loss: 1450.0566
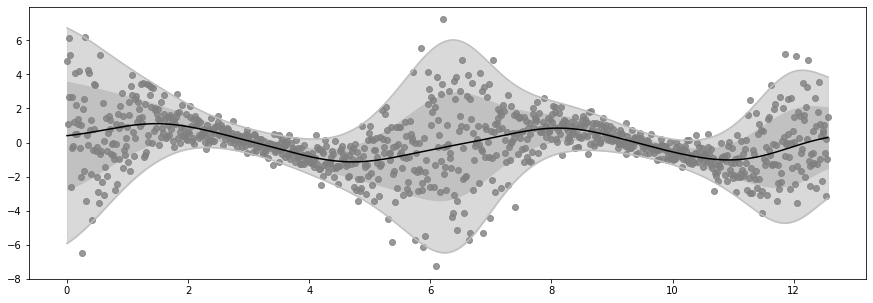
Epoch 100 - Loss: 1449.6072
[9]:
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| SVGP.kernel.kernels[0].variance | Parameter | Softplus | True | () | float64 | 0.8370709041693771 | |
| SVGP.kernel.kernels[0].lengthscales | Parameter | Softplus | True | () | float64 | 1.1847658388491142 | |
| SVGP.kernel.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1.1310082794009104 | |
| SVGP.kernel.kernels[1].lengthscales | Parameter | Softplus | True | () | float64 | 1.0598457971163158 | |
| SVGP.inducing_variable.inducing_variable_list[0].Z | Parameter | Identity | True | (20, 1) | float64 | [[-0.11665065... | |
| SVGP.inducing_variable.inducing_variable_list[1].Z | Parameter | Identity | True | (20, 1) | float64 | [[-0.26968518... | |
| SVGP.q_mu | Parameter | Identity | False | (20, 2) | float64 | [[0.38260666, 1.07056153... | |
| SVGP.q_sqrt | Parameter | FillTriangular | False | (2, 20, 20) | float64 | [[[4.64009332e-01, 0.00000000e+00, 0.00000000e+00... |
Further reading#
See Chained Gaussian Processes by Saul et al. (AISTATS 2016).