Bayesian Gaussian process latent variable model (Bayesian GPLVM)#
This notebook shows how to use the Bayesian GPLVM model. This is an unsupervised learning method usually used for dimensionality reduction. For an in-depth overview of GPLVMs,see [1, 2].
[1]:
import gpflow
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
import gpflow
from gpflow.utilities import ops, print_summary
from gpflow.config import set_default_float, default_float, set_default_summary_fmt
from gpflow.ci_utils import ci_niter
set_default_float(np.float64)
set_default_summary_fmt("notebook")
%matplotlib inline
2022-05-10 11:05:44.178253: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-05-10 11:05:44.178283: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
Data#
We are using the “three phase oil flow” dataset used initially for demonstrating the Generative Topographic mapping from [3].
[2]:
data = np.load("./data/three_phase_oil_flow.npz")
Following the GPflow notation we assume this dataset has a shape of [num_data, output_dim]
[3]:
Y = tf.convert_to_tensor(data["Y"], dtype=default_float())
2022-05-10 11:05:47.090113: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-05-10 11:05:47.090146: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2022-05-10 11:05:47.090165: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (49c966262641): /proc/driver/nvidia/version does not exist
2022-05-10 11:05:47.090426: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
Integer in \([0, 2]\) indicating to which class the data point belongs (shape [num_data,]). Not used for model fitting, only for plotting afterwards.
[4]:
labels = tf.convert_to_tensor(data["labels"])
[5]:
print("Number of points: {} and Number of dimensions: {}".format(Y.shape[0], Y.shape[1]))
Number of points: 100 and Number of dimensions: 12
Model construction#
We start by initializing the required variables:
[6]:
latent_dim = 2 # number of latent dimensions
num_inducing = 20 # number of inducing pts
num_data = Y.shape[0] # number of data points
Initialize via PCA:
[7]:
X_mean_init = ops.pca_reduce(Y, latent_dim)
X_var_init = tf.ones((num_data, latent_dim), dtype=default_float())
Pick inducing inputs randomly from dataset initialization:
[8]:
np.random.seed(1) # for reproducibility
inducing_variable = tf.convert_to_tensor(
np.random.permutation(X_mean_init.numpy())[:num_inducing], dtype=default_float()
)
We construct a Squared Exponential (SE) kernel operating on the two-dimensional latent space. The ARD parameter stands for Automatic Relevance Determination, which in practice means that we learn a different lengthscale for each of the input dimensions. See Manipulating kernels for more information.
[9]:
lengthscales = tf.convert_to_tensor([1.0] * latent_dim, dtype=default_float())
kernel = gpflow.kernels.RBF(lengthscales=lengthscales)
We have all the necessary ingredients to construct the model. GPflow contains an implementation of the Bayesian GPLVM:
[10]:
gplvm = gpflow.models.BayesianGPLVM(
Y,
X_data_mean=X_mean_init,
X_data_var=X_var_init,
kernel=kernel,
inducing_variable=inducing_variable,
)
# Instead of passing an inducing_variable directly, we can also set the num_inducing_variables argument to an integer, which will randomly pick from the data.
We change the default likelihood variance, which is 1, to 0.01.
[11]:
gplvm.likelihood.variance.assign(0.01)
[11]:
<tf.Variable 'UnreadVariable' shape=() dtype=float64, numpy=-4.600266525158521>
Next we optimize the created model. Given that this model has a deterministic evidence lower bound (ELBO), we can use SciPy’s BFGS optimizer.
[12]:
opt = gpflow.optimizers.Scipy()
maxiter = ci_niter(1000)
_ = opt.minimize(
gplvm.training_loss,
method="BFGS",
variables=gplvm.trainable_variables,
options=dict(maxiter=maxiter),
)
2022-05-10 11:05:47.194015: W tensorflow/python/util/util.cc:368] Sets are not currently considered sequences, but this may change in the future, so consider avoiding using them.
Model analysis#
GPflow allows you to inspect the learned model hyperparameters.
[13]:
print_summary(gplvm)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| BayesianGPLVM.kernel.variance | Parameter | Softplus | True | () | float64 | 0.9180117032573774 | |
| BayesianGPLVM.kernel.lengthscales | Parameter | Softplus | True | (2,) | float64 | [0.86661779 1.76000883] | |
| BayesianGPLVM.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.006477223181646814 | |
| BayesianGPLVM.X_data_mean | Parameter | Identity | True | (100, 2) | float64 | [[-7.98770228e-01, 3.04428556e+00... | |
| BayesianGPLVM.X_data_var | Parameter | Softplus | True | (100, 2) | float64 | [[0.00040639, 0.00153663... | |
| BayesianGPLVM.inducing_variable.Z | Parameter | Identity | True | (20, 2) | float64 | [[1.31767770e+00, -1.72368614e+00... |
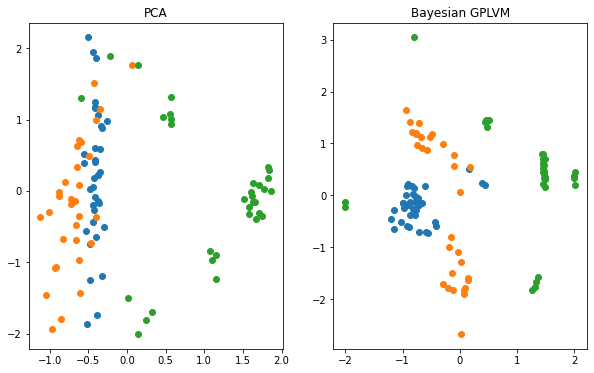
Plotting vs. Principle Component Analysis (PCA)#
The reduction of the dimensionality of the dataset to two dimensions allows us to visualize the learned manifold. We compare the Bayesian GPLVM’s latent space to the deterministic PCA’s one.
[14]:
X_pca = ops.pca_reduce(Y, latent_dim).numpy()
gplvm_X_mean = gplvm.X_data_mean.numpy()
f, ax = plt.subplots(1, 2, figsize=(10, 6))
for i in np.unique(labels):
ax[0].scatter(X_pca[labels == i, 0], X_pca[labels == i, 1], label=i)
ax[1].scatter(gplvm_X_mean[labels == i, 0], gplvm_X_mean[labels == i, 1], label=i)
ax[0].set_title("PCA")
ax[1].set_title("Bayesian GPLVM")
[ ]:
References#
[1] Lawrence, Neil D. ‘Gaussian process latent variable models for visualization of high dimensional data’. Advances in Neural Information Processing Systems. 2004.
[2] Titsias, Michalis, and Neil D. Lawrence. ‘Bayesian Gaussian process latent variable model’. Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics. 2010.
[3] Bishop, Christopher M., and Gwilym D. James. ‘Analysis of multiphase flows using dual-energy gamma densitometry and neural networks’. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 327.2-3 (1993): 580-593.