Comparing FITC approximation to VFE approximation#
This notebook examines why we prefer the Variational Free Energy (VFE) objective to the Fully Independent Training Conditional (FITC) approximation for our sparse approximations.
[1]:
import gpflow
import tensorflow as tf
from gpflow.ci_utils import ci_niter # to speed up automated testing of this notebook
import matplotlib.pyplot as plt
%matplotlib inline
from FITCvsVFE import (
getTrainingTestData,
printModelParameters,
plotPredictions,
repeatMinimization,
stretch,
plotComparisonFigure,
)
import logging
# logging.disable(logging.WARN) # do not clutter up the notebook with optimization warnings
2022-05-10 11:26:23.547549: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-05-10 11:26:23.547576: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
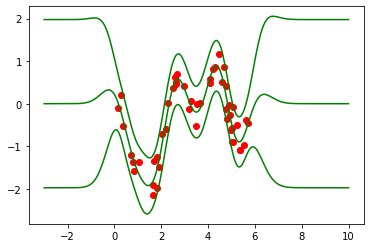
First, we load the training data and plot it together with the exact GP solution (using the GPR model):
[2]:
# Load the training data:
Xtrain, Ytrain, Xtest, Ytest = getTrainingTestData()
def getKernel():
return gpflow.kernels.SquaredExponential()
# Run exact inference on training data:
exact_model = gpflow.models.GPR((Xtrain, Ytrain), kernel=getKernel())
opt = gpflow.optimizers.Scipy()
opt.minimize(
exact_model.training_loss,
exact_model.trainable_variables,
method="L-BFGS-B",
options=dict(maxiter=ci_niter(20000)),
tol=1e-11,
)
print("Exact model parameters:")
printModelParameters(exact_model)
figA, ax = plt.subplots(1, 1)
ax.plot(Xtrain, Ytrain, "ro")
plotPredictions(ax, exact_model, color="g")
2022-05-10 11:26:26.491193: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-05-10 11:26:26.491225: W tensorflow/stream_executor/cuda/cuda_driver.cc:269] failed call to cuInit: UNKNOWN ERROR (303)
2022-05-10 11:26:26.491242: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (49c966262641): /proc/driver/nvidia/version does not exist
2022-05-10 11:26:26.491504: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-05-10 11:26:26.522050: W tensorflow/python/util/util.cc:368] Sets are not currently considered sequences, but this may change in the future, so consider avoiding using them.
Exact model parameters:
Likelihood variance = 0.074285
Kernel variance = 0.90049
Kernel lengthscale = 0.5825
[3]:
def initializeHyperparametersFromExactSolution(sparse_model):
sparse_model.likelihood.variance.assign(exact_model.likelihood.variance)
sparse_model.kernel.variance.assign(exact_model.kernel.variance)
sparse_model.kernel.lengthscales.assign(exact_model.kernel.lengthscales)
We now construct two sparse model using the VFE (SGPR model) and FITC (GPRFITC model) optimization objectives, with the inducing points being initialized on top of the training inputs, and the model hyperparameters (kernel variance and lengthscales, and likelihood variance) being initialized to the values obtained in the optimization of the exact GPR model:
[4]:
# Train VFE model initialized from the perfect solution.
VFEmodel = gpflow.models.SGPR((Xtrain, Ytrain), kernel=getKernel(), inducing_variable=Xtrain.copy())
initializeHyperparametersFromExactSolution(VFEmodel)
VFEcb = repeatMinimization(VFEmodel, Xtest, Ytest) # optimize with several restarts
print("Sparse model parameters after VFE optimization:")
printModelParameters(VFEmodel)
WARNING:tensorflow:5 out of the last 14 calls to <function Scipy.eval_func.<locals>._tf_eval at 0x7f919426fac0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
WARNING:tensorflow:5 out of the last 14 calls to <function Scipy.eval_func.<locals>._tf_eval at 0x7f91647af520> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
Sparse model parameters after VFE optimization:
Likelihood variance = 0.074286
Kernel variance = 0.90049
Kernel lengthscale = 0.5825
[5]:
# Train FITC model initialized from the perfect solution.
FITCmodel = gpflow.models.GPRFITC(
(Xtrain, Ytrain), kernel=getKernel(), inducing_variable=Xtrain.copy()
)
initializeHyperparametersFromExactSolution(FITCmodel)
FITCcb = repeatMinimization(FITCmodel, Xtest, Ytest) # optimize with several restarts
print("Sparse model parameters after FITC optimization:")
printModelParameters(FITCmodel)
Sparse model parameters after FITC optimization:
Likelihood variance = 0.018908
Kernel variance = 1.3343
Kernel lengthscale = 0.61751
Plotting a comparison of the two algorithms, we see that VFE stays at the optimum of exact GPR, whereas the FITC approximation eventually ends up with several inducing points on top of each other, and a worse fit:
[6]:
figB, axes = plt.subplots(3, 2, figsize=(20, 16))
# VFE optimization finishes after 10 iterations, so we stretch out the training and test
# log-likelihood traces to make them comparable against FITC:
VFEiters = FITCcb.n_iters
VFElog_likelihoods = stretch(len(VFEiters), VFEcb.log_likelihoods)
VFEhold_out_likelihood = stretch(len(VFEiters), VFEcb.hold_out_likelihood)
axes[0, 0].set_title("VFE", loc="center", fontdict={"fontsize": 22})
plotComparisonFigure(
Xtrain,
VFEmodel,
exact_model,
axes[0, 0],
axes[1, 0],
axes[2, 0],
VFEiters,
VFElog_likelihoods,
VFEhold_out_likelihood,
)
axes[0, 1].set_title("FITC", loc="center", fontdict={"fontsize": 22})
plotComparisonFigure(
Xtrain,
FITCmodel,
exact_model,
axes[0, 1],
axes[1, 1],
axes[2, 1],
FITCcb.n_iters,
FITCcb.log_likelihoods,
FITCcb.hold_out_likelihood,
)
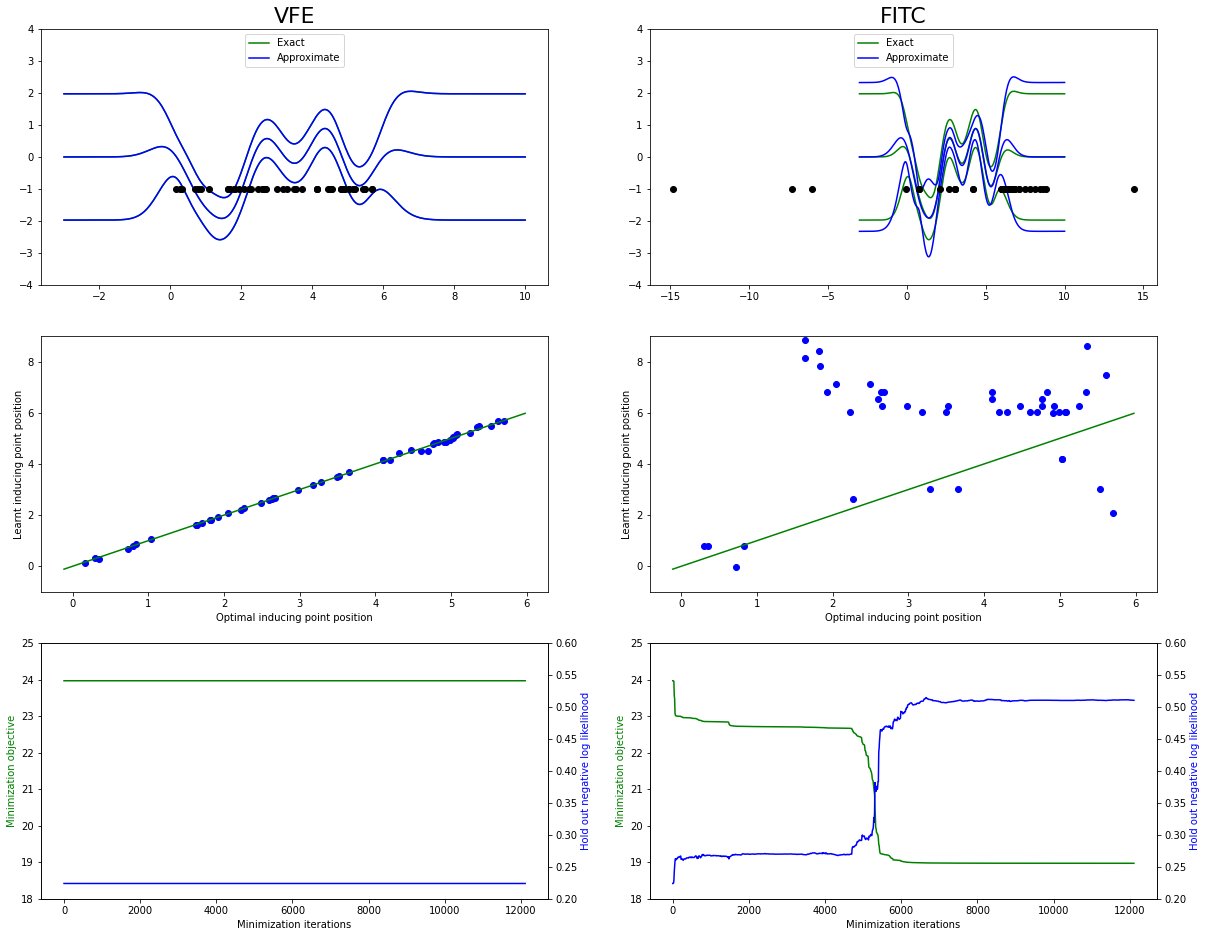
A more detailed discussion of the comparison between these sparse approximations can be found in Understanding Probabilistic Sparse Gaussian Process Approximations by Bauer, van der Wilk, and Rasmussen (2017).