A simple demonstration of coregionalization#
This notebook shows how to construct a multi-output GP model using GPflow. We will consider a regression problem for functions \(f: \mathbb{R}^D \rightarrow \mathbb{R}^P\). We assume that the dataset is of the form \((X_1, f_1), \dots, (X_P, f_P)\), that is, we do not necessarily observe all the outputs for a particular input location (for cases where there are fully observed outputs for each input, see Multi-output Gaussian processes in GPflow for a more efficient implementation). We allow each \(f_i\) to have a different noise distribution by assigning a different likelihood to each.
For this problem, we model \(f\) as a coregionalized Gaussian process, which assumes a kernel of the form:
\begin{equation} \textrm{cov}(f_i(X), f_j(X^\prime)) = k(X, X^\prime) \cdot B[i, j]. \end{equation}
The covariance of the \(i\)th function at \(X\) and the \(j\)th function at \(X^\prime\) is a kernel applied at \(X\) and \(X^\prime\), times the \((i, j)\)th entry of a positive definite \(P \times P\) matrix \(B\). This is known as the intrinsic model of coregionalization (ICM) (Bonilla and Williams, 2008).
To make sure that B is positive-definite, we parameterize it as:
\begin{equation} B = W W^\top + \textrm{diag}(\kappa). \end{equation}
To build such a model in GPflow, we need to perform the following two steps:
Create the kernel function defined previously, using the
Coregionkernel class.Augment the training data X with an extra column that contains an integer index to indicate which output an observation is associated with. This is essential to make the data work with the
Coregionkernel.Create a likelihood for each output using the
SwitchedLikelihoodclass, which is a container for other likelihoods.Augment the training data Y with an extra column that contains an integer index to indicate which likelihood an observation is associated with.
[1]:
import gpflow
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from gpflow.ci_utils import reduce_in_tests
plt.rcParams["figure.figsize"] = (12, 6)
np.random.seed(123)
2022-09-20 14:23:35.960378: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-09-20 14:23:36.111878: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-09-20 14:23:36.111900: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
2022-09-20 14:23:36.144135: E tensorflow/stream_executor/cuda/cuda_blas.cc:2981] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
2022-09-20 14:23:37.092009: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory
2022-09-20 14:23:37.092079: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory
2022-09-20 14:23:37.092087: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.decorator.check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.inheritance.inherit_check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
Data preparation#
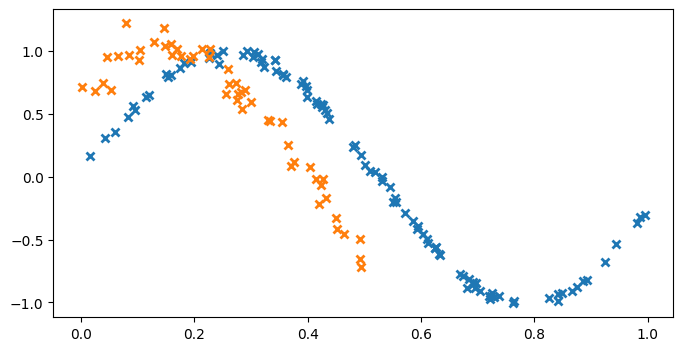
We start by generating some training data to fit the model with. For this example, we choose the following two correlated functions for our outputs:
\begin{align} y_1 &= \sin(6x) + \epsilon_1, \qquad \epsilon_1 \sim \mathcal{N}(0, 0.009) \\ y_2 &= \sin(6x + 0.7) + \epsilon_2, \qquad \epsilon_2 \sim \mathcal{N}(0, 0.01) \end{align}
[2]:
# make a dataset with two outputs, correlated, heavy-tail noise. One has more noise than the other.
X1 = np.random.rand(100, 1) # Observed locations for first output
X2 = np.random.rand(50, 1) * 0.5 # Observed locations for second output
Y1 = np.sin(6 * X1) + np.random.randn(*X1.shape) * 0.03
Y2 = np.sin(6 * X2 + 0.7) + np.random.randn(*X2.shape) * 0.1
plt.figure(figsize=(8, 4))
plt.plot(X1, Y1, "x", mew=2)
_ = plt.plot(X2, Y2, "x", mew=2)
Data formatting for the coregionalized model#
We add an extra column to our training dataset that contains an index that specifies which output is observed.
[3]:
# Augment the input with ones or zeros to indicate the required output dimension
X_augmented = np.vstack((np.hstack((X1, np.zeros_like(X1))), np.hstack((X2, np.ones_like(X2)))))
# Augment the Y data with ones or zeros that specify a likelihood from the list of likelihoods
Y_augmented = np.vstack((np.hstack((Y1, np.zeros_like(Y1))), np.hstack((Y2, np.ones_like(Y2)))))
Building the coregionalization kernel#
We build a coregionalization kernel with the Matern 3/2 kernel as the base kernel. This acts on the leading ([0]) data dimension of the augmented X values. The Coregion kernel indexes the outputs, and acts on the last ([1]) data dimension (indices) of the augmented X values. To specify these dimensions, we use the built-in active_dims argument in the kernel constructor. To construct the full multi-output kernel, we take the product of the two kernels (for a more in-depth tutorial on
kernel combination, see Manipulating kernels).
[4]:
output_dim = 2 # Number of outputs
rank = 1 # Rank of W
# Base kernel
k = gpflow.kernels.Matern32(active_dims=[0])
# Coregion kernel
coreg = gpflow.kernels.Coregion(output_dim=output_dim, rank=rank, active_dims=[1])
kern = k * coreg
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.checker.ShapeChecker.__init__ which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
2022-09-20 14:23:40.683283: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-09-20 14:23:40.683754: W tensorflow/stream_executor/cuda/cuda_driver.cc:263] failed call to cuInit: UNKNOWN ERROR (303)
2022-09-20 14:23:40.683778: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (947c84c595a3): /proc/driver/nvidia/version does not exist
2022-09-20 14:23:40.684529: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
Note: W = 0 is a saddle point in the objective, which would result in the value of W not being optimized to fit the data. Hence, by default, the W matrix is initialized with 0.1. Alternatively, you could re-initialize the matrix to random entries.
Constructing the model#
The final element in building the model is to specify the likelihood for each output dimension. To do this, use the SwitchedLikelihood object in GPflow.
[5]:
# This likelihood switches between Gaussian noise with different variances for each f_i:
lik = gpflow.likelihoods.SwitchedLikelihood(
[gpflow.likelihoods.Gaussian(), gpflow.likelihoods.Gaussian()]
)
# now build the GP model as normal
m = gpflow.models.VGP((X_augmented, Y_augmented), kernel=kern, likelihood=lik)
# fit the covariance function parameters
maxiter = reduce_in_tests(10000)
gpflow.optimizers.Scipy().minimize(
m.training_loss,
m.trainable_variables,
options=dict(maxiter=maxiter),
method="L-BFGS-B",
)
[5]:
fun: -225.66181154535906
hess_inv: <11483x11483 LbfgsInvHessProduct with dtype=float64>
jac: array([-3.36694015e-01, -2.45082430e-02, 7.13931131e-04, ...,
-2.88453100e-04, -3.25729946e-03, -2.60972009e-04])
message: 'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'
nfev: 1519
nit: 1378
njev: 1519
status: 0
success: True
x: array([-0.79481659, 2.21464663, -1.35600031, ..., -1.79021103,
-7.38931801, -4.8347069 ])
That’s it: the model is trained. Let’s plot the model fit to see what’s happened.
[6]:
def plot_gp(x, mu, var, color, label):
plt.plot(x, mu, color=color, lw=2, label=label)
plt.fill_between(
x[:, 0],
(mu - 2 * np.sqrt(var))[:, 0],
(mu + 2 * np.sqrt(var))[:, 0],
color=color,
alpha=0.4,
)
def plot(m):
plt.figure(figsize=(8, 4))
Xtest = np.linspace(0, 1, 100)[:, None]
(line,) = plt.plot(X1, Y1, "x", mew=2)
mu, var = m.predict_f(np.hstack((Xtest, np.zeros_like(Xtest))))
plot_gp(Xtest, mu, var, line.get_color(), "Y1")
(line,) = plt.plot(X2, Y2, "x", mew=2)
mu, var = m.predict_f(np.hstack((Xtest, np.ones_like(Xtest))))
plot_gp(Xtest, mu, var, line.get_color(), "Y2")
plt.legend()
plot(m)
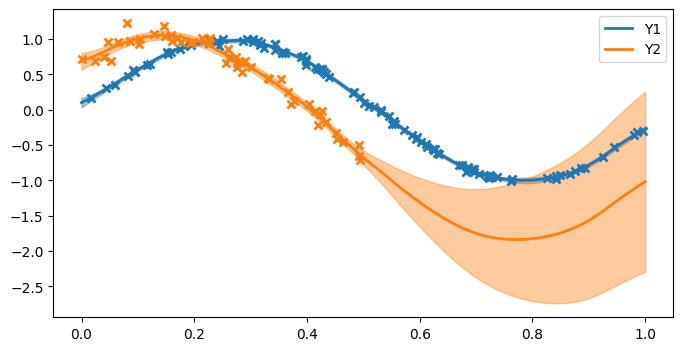
From the plots, we can see:
The first function (blue) has low posterior variance everywhere because there are so many observations, and the noise variance is small.
The second function (orange) has higher posterior variance near the data, because the data are more noisy, and very high posterior variance where there are no observations (x > 0.5).
The model has done a reasonable job of estimating the noise variance and lengthscale.
The model recognises the correlation between the two functions and is able to suggest (with uncertainty) that because x > 0.5 the orange curve should follow the blue curve (which we know to be the truth from the data-generating procedure).
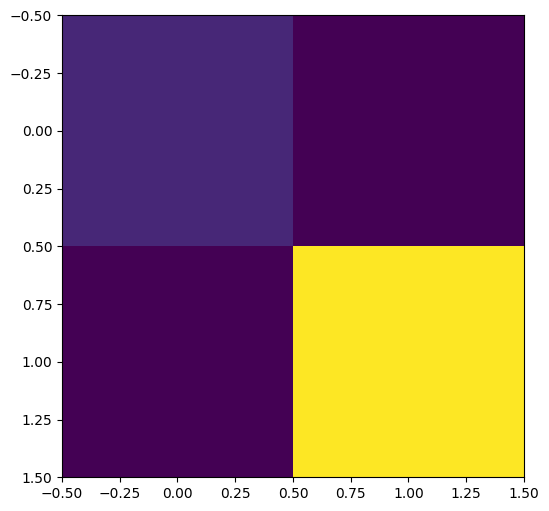
The covariance matrix between outputs is as follows:
[7]:
B = coreg.output_covariance().numpy()
print("B =", B)
_ = plt.imshow(B)
B = [[2.85387406 2.65422127]
[2.65422127 4.44798141]]
References#
Bonilla, Edwin V., Kian M. Chai, and Christopher Williams. “Multi-task Gaussian process prediction.” Advances in neural information processing systems. 2008.