Manipulating kernels#
GPflow comes with a range of kernels. In this notebook, we examine some of them, show how you can combine them to make new kernels, and discuss the active_dims feature.
[1]:
import gpflow
import numpy as np
import matplotlib.pyplot as plt
plt.style.use("ggplot")
import tensorflow as tf
from gpflow.ci_utils import reduce_in_tests
%matplotlib inline
2022-09-20 14:23:32.389983: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-09-20 14:23:32.513878: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-09-20 14:23:32.513901: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
2022-09-20 14:23:32.541884: E tensorflow/stream_executor/cuda/cuda_blas.cc:2981] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
2022-09-20 14:23:33.264285: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory
2022-09-20 14:23:33.264348: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory
2022-09-20 14:23:33.264356: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.decorator.check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.inheritance.inherit_check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
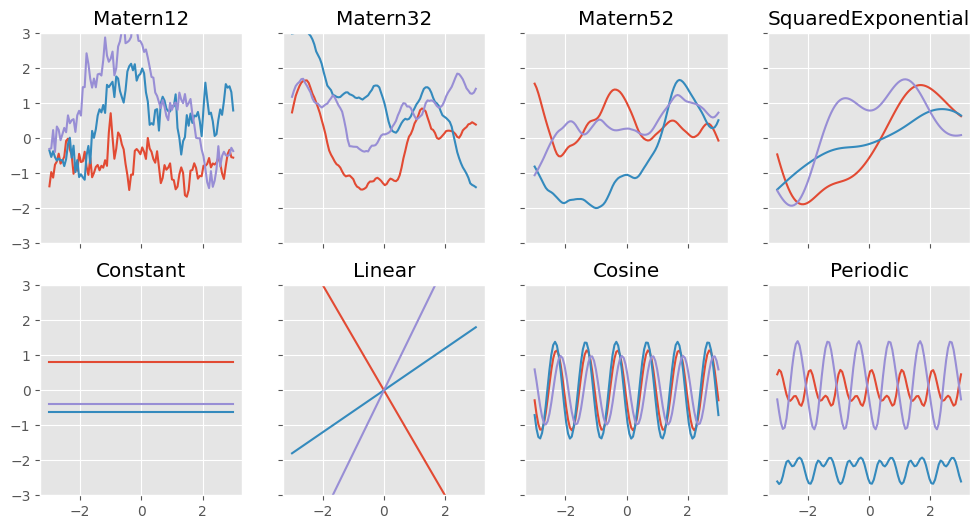
Standard kernels in GPflow#
GPflow comes with lots of standard kernels. Some very simple kernels produce constant functions, linear functions, and white noise functions:
gpflow.kernels.Constantgpflow.kernels.Lineargpflow.kernels.White
Some stationary functions produce samples with varying degrees of smoothness:
gpflow.kernels.Exponentialgpflow.kernels.Matern12gpflow.kernels.Matern32gpflow.kernels.Matern52gpflow.kernels.SquaredExponential(also known asgpflow.kernels.RBF)gpflow.kernels.RationalQuadratic
Two kernels produce periodic samples:
gpflow.kernels.Cosinegpflow.kernels.Periodic
Other kernels that are implemented in core GPflow include:
gpflow.kernels.Polynomialgpflow.kernels.ArcCosine(“neural network kernel”)gpflow.kernels.Coregion
Let’s define some plotting utils functions and have a look at samples from the prior for some of them:
[2]:
def plotkernelsample(k, ax, xmin=-3, xmax=3):
n_grid = reduce_in_tests(100)
xx = np.linspace(xmin, xmax, n_grid)[:, None]
K = k(xx)
ax.plot(xx, np.random.multivariate_normal(np.zeros(n_grid), K, 3).T)
ax.set_title(k.__class__.__name__)
np.random.seed(27)
f, axes = plt.subplots(2, 4, figsize=(12, 6), sharex=True, sharey=True)
plotkernelsample(gpflow.kernels.Matern12(), axes[0, 0])
plotkernelsample(gpflow.kernels.Matern32(), axes[0, 1])
plotkernelsample(gpflow.kernels.Matern52(), axes[0, 2])
plotkernelsample(gpflow.kernels.RBF(), axes[0, 3])
plotkernelsample(gpflow.kernels.Constant(), axes[1, 0])
plotkernelsample(gpflow.kernels.Linear(), axes[1, 1])
plotkernelsample(gpflow.kernels.Cosine(), axes[1, 2])
plotkernelsample(gpflow.kernels.Periodic(gpflow.kernels.SquaredExponential()), axes[1, 3])
_ = axes[0, 0].set_ylim(-3, 3)
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.checker.ShapeChecker.__init__ which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
2022-09-20 14:23:36.473127: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-09-20 14:23:36.473522: W tensorflow/stream_executor/cuda/cuda_driver.cc:263] failed call to cuInit: UNKNOWN ERROR (303)
2022-09-20 14:23:36.473544: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (8b64c123e584): /proc/driver/nvidia/version does not exist
2022-09-20 14:23:36.474298: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
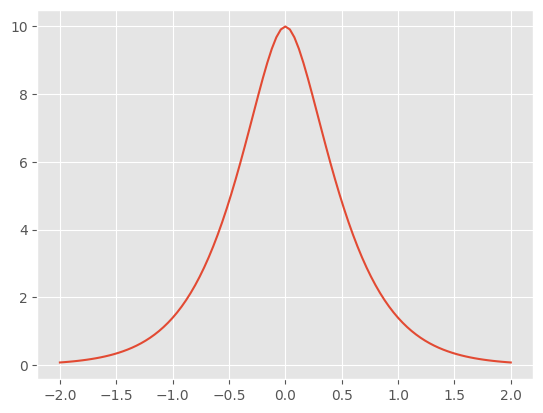
First example: create a Matern 3/2 covariance kernel#
Many kernels have hyperparameters, for example variance and lengthscales. You can change the value of these parameters from their default value of 1.0.
[3]:
k = gpflow.kernels.Matern32(variance=10.0, lengthscales=2)
NOTE: The values specified for the variance and lengthscales parameters are floats.
To get information about the kernel, use print_summary(k) (plain text) or, in a notebook, pass the option fmt="notebook" to obtain a nicer rendering:
[4]:
from gpflow.utilities import print_summary
print_summary(k)
print_summary(k, fmt="notebook")
# You can change the default format as follows:
gpflow.config.set_default_summary_fmt("notebook")
print_summary(k)
╒═══════════════════════╤═══════════╤═════════════╤═════════╤═════════════╤═════════╤═════════╤═════════╕
│ name │ class │ transform │ prior │ trainable │ shape │ dtype │ value │
╞═══════════════════════╪═══════════╪═════════════╪═════════╪═════════════╪═════════╪═════════╪═════════╡
│ Matern32.variance │ Parameter │ Softplus │ │ True │ () │ float64 │ 10 │
├───────────────────────┼───────────┼─────────────┼─────────┼─────────────┼─────────┼─────────┼─────────┤
│ Matern32.lengthscales │ Parameter │ Softplus │ │ True │ () │ float64 │ 2 │
╘═══════════════════════╧═══════════╧═════════════╧═════════╧═════════════╧═════════╧═════════╧═════════╛
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Matern32.variance | Parameter | Softplus | True | () | float64 | 10 | |
| Matern32.lengthscales | Parameter | Softplus | True | () | float64 | 2 |
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Matern32.variance | Parameter | Softplus | True | () | float64 | 10 | |
| Matern32.lengthscales | Parameter | Softplus | True | () | float64 | 2 |
You can access the parameter values and assign new values with the same syntax as for models:
[5]:
print(k.lengthscales)
k.lengthscales.assign(0.5)
print(k.lengthscales)
<Parameter: name=softplus, dtype=float64, shape=[], fn="softplus", numpy=2.0>
<Parameter: name=softplus, dtype=float64, shape=[], fn="softplus", numpy=0.5>
Finally, you can call the kernel object to compute covariance matrices:
[6]:
X1 = np.array([[0.0]])
X2 = np.linspace(-2, 2, 101).reshape(-1, 1)
K21 = k(X2, X1) # cov(f(X2), f(X1)): matrix with shape [101, 1]
K22 = k(X2) # equivalent to k(X2, X2) (but more efficient): matrix with shape [101, 101]
# plotting
plt.figure()
_ = plt.plot(X2, K21)
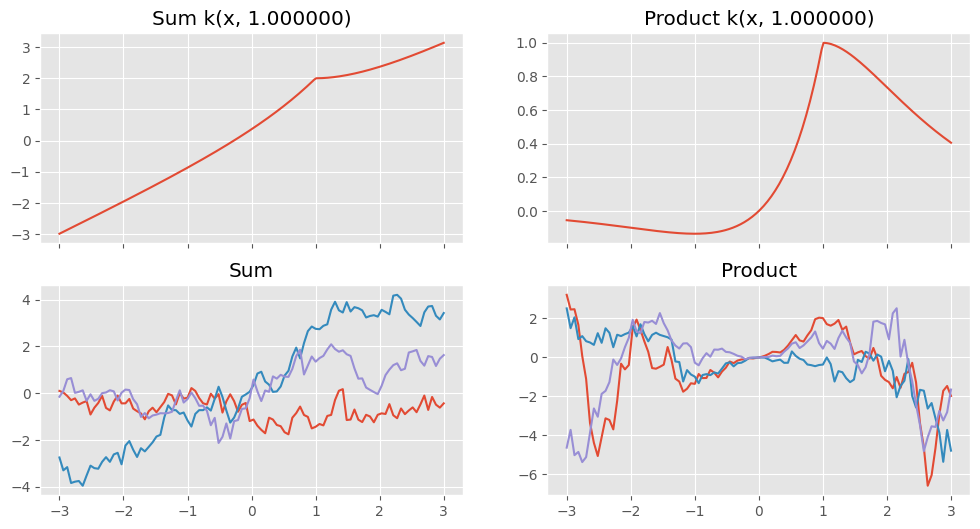
Combine kernels#
Sums and products of kernels are also valid kernels. You can add or multiply instances of kernels to create a new composite kernel with the parameters of the old ones:
[7]:
k1 = gpflow.kernels.Matern12()
k2 = gpflow.kernels.Linear()
k3 = k1 + k2
k4 = k1 * k2
print_summary(k3)
print_summary(k4)
def plotkernelfunction(k, ax, xmin=-3, xmax=3, other=0):
n_grid = reduce_in_tests(200)
xx = np.linspace(xmin, xmax, n_grid)[:, None]
ax.plot(xx, k(xx, np.zeros((1, 1)) + other))
ax.set_title(k.__class__.__name__ + " k(x, %f)" % other)
f, axes = plt.subplots(2, 2, figsize=(12, 6), sharex=True)
plotkernelfunction(k3, axes[0, 0], other=1.0)
plotkernelfunction(k4, axes[0, 1], other=1.0)
plotkernelsample(k3, axes[1, 0])
plotkernelsample(k4, axes[1, 1])
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Sum.kernels[0].variance | Parameter | Softplus | True | () | float64 | 1 | |
| Sum.kernels[0].lengthscales | Parameter | Softplus | True | () | float64 | 1 | |
| Sum.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1 |
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Product.kernels[0].variance | Parameter | Softplus | True | () | float64 | 1 | |
| Product.kernels[0].lengthscales | Parameter | Softplus | True | () | float64 | 1 | |
| Product.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1 |
Kernels for higher-dimensional input spaces#
Kernels generalize to multiple dimensions straightforwardly. Stationary kernels support “Automatic Relevance Determination” (ARD), that is, having a different lengthscale parameter for each input dimension. Simply pass in an array of the same length as the number of input dimensions. NOTE: This means that the kernel object is then able to process only inputs of that dimension!
You can also initialize the lengthscales when the object is created:
[8]:
k = gpflow.kernels.Matern52(lengthscales=[0.1, 0.2, 5.0])
print_summary(k)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Matern52.variance | Parameter | Softplus | True | () | float64 | 1.0 | |
| Matern52.lengthscales | Parameter | Softplus | True | (3,) | float64 | [0.1 0.2 5. ] |
Specify active dimensions#
When combining kernels, it’s often helpful to have bits of the kernel working on different dimensions. For example, to model a function that is linear in the first dimension and smooth in the second, we could use a combination of Linear and Matern52 kernels, one for each dimension.
To tell GPflow which dimension a kernel applies to, specify a list of integers as the value of the active_dims parameter.
[9]:
k1 = gpflow.kernels.Linear(active_dims=[0])
k2 = gpflow.kernels.Matern52(active_dims=[1])
k = k1 + k2
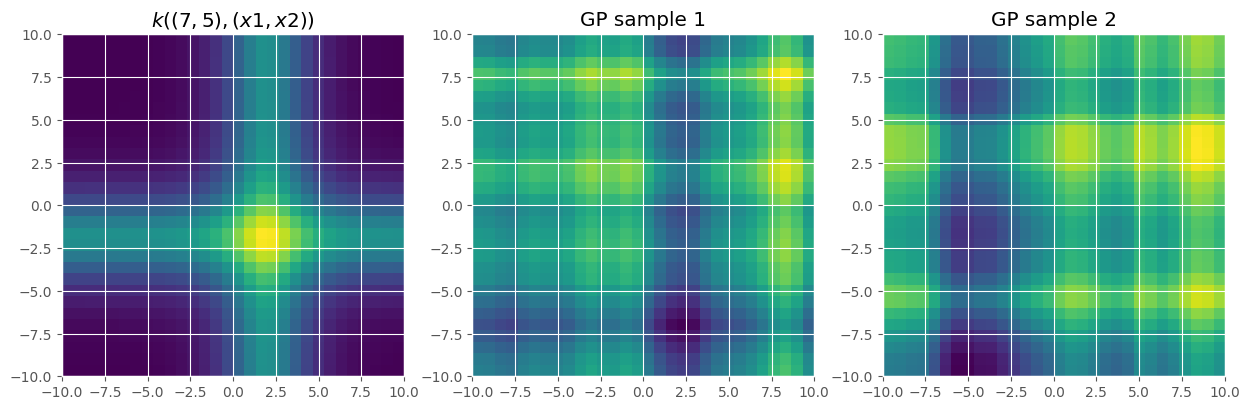
active_dims makes it easy to create additive models. Here we build an additive Matern 5/2 kernel:
[10]:
k = gpflow.kernels.Matern52(active_dims=[0], lengthscales=2) + gpflow.kernels.Matern52(
active_dims=[1], lengthscales=2
)
Let’s plot this kernel and sample from it:
[11]:
n_grid = reduce_in_tests(30)
x = np.linspace(-10, 10, n_grid)
X, Y = np.meshgrid(x, x)
X = np.vstack((X.flatten(), Y.flatten())).T
x0 = np.array([[2.0, 2.0]])
# plot the kernel
KxX = k(X, x0).numpy().reshape(n_grid, n_grid)
fig, axes = plt.subplots(1, 3, figsize=(15, 5))
axes[0].imshow(KxX, extent=[-10, 10, -10, 10])
axes[0].set_title(f"$k((7, 5), (x1, x2))$")
# plot a GP sample
K = k(X).numpy()
Z = np.random.multivariate_normal(np.zeros(n_grid ** 2), K, 2)
axes[1].imshow(Z[0, :].reshape(n_grid, n_grid), extent=[-10, 10, -10, 10])
axes[1].set_title("GP sample 1")
axes[2].imshow(Z[1, :].reshape(n_grid, n_grid), extent=[-10, 10, -10, 10])
_ = axes[2].set_title("GP sample 2")
Define new covariance functions#
GPflow makes it easy to define new covariance functions. See Kernel design for more information.