Basic (Gaussian likelihood) GP regression model#
This notebook shows the different steps for creating and using a standard GP regression model, including: - reading and formatting data - choosing a kernel function - choosing a mean function (optional) - creating the model - viewing, getting, and setting model parameters - optimizing the model parameters - making predictions
We focus here on the implementation of the models in GPflow; for more intuition on these models, see A Practical Guide to Gaussian Processes and A Visual Exploration of Gaussian Processes.
[1]:
import gpflow
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from gpflow.utilities import print_summary
# The lines below are specific to the notebook format
%matplotlib inline
plt.rcParams["figure.figsize"] = (12, 6)
2022-09-20 14:25:37.380602: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-09-20 14:25:37.518073: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-09-20 14:25:37.518093: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
2022-09-20 14:25:37.545914: E tensorflow/stream_executor/cuda/cuda_blas.cc:2981] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
2022-09-20 14:25:38.370737: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory
2022-09-20 14:25:38.370804: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory
2022-09-20 14:25:38.370812: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.decorator.check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.inheritance.inherit_check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
X and Y denote the input and output values. NOTE: X and Y must be two-dimensional NumPy arrays, \(N \times 1\) or \(N \times D\), where \(D\) is the number of input dimensions/features, with the same number of rows as \(N\) (one for each data point):
[2]:
data = np.genfromtxt("data/regression_1D.csv", delimiter=",")
X = data[:, 0].reshape(-1, 1)
Y = data[:, 1].reshape(-1, 1)
_ = plt.plot(X, Y, "kx", mew=2)
We will consider the following probabilistic model: \begin{equation} Y_i = f(X_i) + \varepsilon_i\,, \end{equation} where \(f \sim \mathcal{GP}(\mu(\cdot), k(\cdot, \cdot'))\), and \(\varepsilon \sim \mathcal{N}(0, \tau^2 I)\).
Choose a kernel#
Several kernels (covariance functions) are implemented in GPflow. You can easily combine them to create new ones (see Manipulating kernels). You can also implement new covariance functions, as shown in the Kernel design notebook. Here, we will use a simple one:
[3]:
k = gpflow.kernels.Matern52()
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.checker.ShapeChecker.__init__ which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
2022-09-20 14:25:41.676195: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-09-20 14:25:41.676222: W tensorflow/stream_executor/cuda/cuda_driver.cc:263] failed call to cuInit: UNKNOWN ERROR (303)
2022-09-20 14:25:41.676240: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (c3ed7e624fe5): /proc/driver/nvidia/version does not exist
2022-09-20 14:25:41.676494: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
For more advanced kernels see the advanced kernel notebook (including kernels defined on subspaces). A summary of the kernel can be obtained by
[4]:
print_summary(k)
╒═══════════════════════╤═══════════╤═════════════╤═════════╤═════════════╤═════════╤═════════╤═════════╕
│ name │ class │ transform │ prior │ trainable │ shape │ dtype │ value │
╞═══════════════════════╪═══════════╪═════════════╪═════════╪═════════════╪═════════╪═════════╪═════════╡
│ Matern52.variance │ Parameter │ Softplus │ │ True │ () │ float64 │ 1 │
├───────────────────────┼───────────┼─────────────┼─────────┼─────────────┼─────────┼─────────┼─────────┤
│ Matern52.lengthscales │ Parameter │ Softplus │ │ True │ () │ float64 │ 1 │
╘═══════════════════════╧═══════════╧═════════════╧═════════╧═════════════╧═════════╧═════════╧═════════╛
The Matern 5/2 kernel has two parameters: lengthscales, which encodes the “wiggliness” of the GP, and variance, which tunes the amplitude. They are both set to 1.0 as the default value. For more details on the meaning of the other columns, see Manipulating kernels.
Choose a mean function (optional)#
It is common to choose \(\mu = 0\), which is the GPflow default.
However, if there is a clear pattern (such as a mean value of Y that is far away from 0, or a linear trend in the data), mean functions can be beneficial. Some simple ones are provided in the gpflow.mean_functions module.
Here’s how to define a linear mean function:
meanf = gpflow.mean_functions.Linear()
Construct a model#
A GPflow model is created by instantiating one of the GPflow model classes, in this case GPR. We’ll make a kernel k and instantiate a GPR object using the generated data and the kernel. We’ll also set the variance of the likelihood to a sensible initial guess.
[5]:
m = gpflow.models.GPR(data=(X, Y), kernel=k, mean_function=None)
A summary of the model can be obtained by
[6]:
print_summary(m)
╒═════════════════════════╤═══════════╤══════════════════╤═════════╤═════════════╤═════════╤═════════╤═════════╕
│ name │ class │ transform │ prior │ trainable │ shape │ dtype │ value │
╞═════════════════════════╪═══════════╪══════════════════╪═════════╪═════════════╪═════════╪═════════╪═════════╡
│ GPR.kernel.variance │ Parameter │ Softplus │ │ True │ () │ float64 │ 1 │
├─────────────────────────┼───────────┼──────────────────┼─────────┼─────────────┼─────────┼─────────┼─────────┤
│ GPR.kernel.lengthscales │ Parameter │ Softplus │ │ True │ () │ float64 │ 1 │
├─────────────────────────┼───────────┼──────────────────┼─────────┼─────────────┼─────────┼─────────┼─────────┤
│ GPR.likelihood.variance │ Parameter │ Softplus + Shift │ │ True │ () │ float64 │ 1 │
╘═════════════════════════╧═══════════╧══════════════════╧═════════╧═════════════╧═════════╧═════════╧═════════╛
The first two lines correspond to the kernel parameters, and the third one gives the likelihood parameter (the noise variance \(\tau^2\) in our model).
You can access those values and manually set them to sensible initial guesses. For example:
[7]:
m.likelihood.variance.assign(0.01)
m.kernel.lengthscales.assign(0.3)
[7]:
<tf.Variable 'UnreadVariable' shape=() dtype=float64, numpy=-1.0502256128148466>
Optimize the model parameters#
To obtain meaningful predictions, you need to tune the model parameters (that is, the parameters of the kernel, the likelihood, and the mean function if applicable) to the data at hand.
There are several optimizers available in GPflow. Here we use the Scipy optimizer, which by default implements the L-BFGS-B algorithm. (You can select other algorithms by using the method= keyword argument to its minimize method; see the SciPy documentation for details of available options.)
[8]:
opt = gpflow.optimizers.Scipy()
In order to train the model, we need to maximize the log marginal likelihood. GPflow models define a training_loss that can be passed to the minimize method of an optimizer; in this case it is simply the negative log marginal likelihood. We also need to specify the variables to train with m.trainable_variables, and the number of iterations.
[9]:
opt_logs = opt.minimize(m.training_loss, m.trainable_variables, options=dict(maxiter=100))
print_summary(m)
╒═════════════════════════╤═══════════╤══════════════════╤═════════╤═════════════╤═════════╤═════════╤═════════╕
│ name │ class │ transform │ prior │ trainable │ shape │ dtype │ value │
╞═════════════════════════╪═══════════╪══════════════════╪═════════╪═════════════╪═════════╪═════════╪═════════╡
│ GPR.kernel.variance │ Parameter │ Softplus │ │ True │ () │ float64 │ 7.96581 │
├─────────────────────────┼───────────┼──────────────────┼─────────┼─────────────┼─────────┼─────────┼─────────┤
│ GPR.kernel.lengthscales │ Parameter │ Softplus │ │ True │ () │ float64 │ 0.21242 │
├─────────────────────────┼───────────┼──────────────────┼─────────┼─────────────┼─────────┼─────────┼─────────┤
│ GPR.likelihood.variance │ Parameter │ Softplus + Shift │ │ True │ () │ float64 │ 0.00576 │
╘═════════════════════════╧═══════════╧══════════════════╧═════════╧═════════════╧═════════╧═════════╧═════════╛
Notice how the value column has changed.
The local optimum found by Maximum Likelihood might not be the one you want (for example, it might be overfitting or oversmooth). This depends on the initial values of the hyperparameters, and is specific to each dataset. As an alternative to Maximum Likelihood, Markov Chain Monte Carlo (MCMC) is also available.
Make predictions#
We can now use the model to make some predictions at the new points Xnew. You might be interested in predicting two different quantities: the latent function values f(Xnew) (the denoised signal), or the values of new observations y(Xnew) (signal + noise). Because we are dealing with Gaussian probabilistic models, the predictions typically produce a mean and variance as output. Alternatively, you can obtain samples of f(Xnew) or the log density of the new data points
(Xnew, Ynew).
GPflow models have several prediction methods:
m.predict_freturns the mean and marginal variance of \(f\) at the pointsXnew.m.predict_fwith argumentfull_cov=Truereturns the mean and the full covariance matrix of \(f\) at the pointsXnew.m.predict_f_samplesreturns samples of the latent function.m.predict_yreturns the mean and variance of a new data point (that is, it includes the noise variance).m.predict_log_densityreturns the log density of the observationsYnewatXnew.
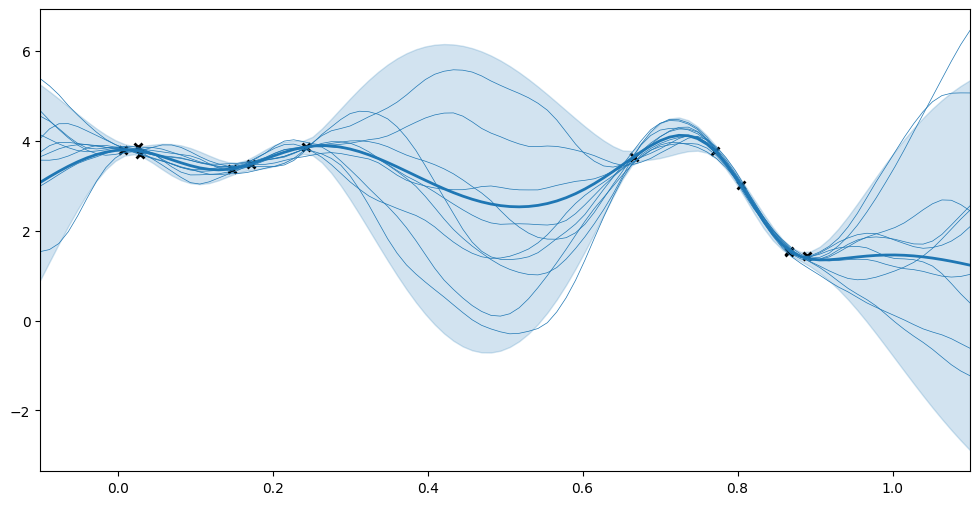
We use predict_f and predict_f_samples to plot 95% confidence intervals and samples from the posterior distribution.
[10]:
## generate test points for prediction
xx = np.linspace(-0.1, 1.1, 100).reshape(100, 1) # test points must be of shape (N, D)
## predict mean and variance of latent GP at test points
mean, var = m.predict_f(xx)
## generate 10 samples from posterior
tf.random.set_seed(1) # for reproducibility
samples = m.predict_f_samples(xx, 10) # shape (10, 100, 1)
## plot
plt.figure(figsize=(12, 6))
plt.plot(X, Y, "kx", mew=2)
plt.plot(xx, mean, "C0", lw=2)
plt.fill_between(
xx[:, 0],
mean[:, 0] - 1.96 * np.sqrt(var[:, 0]),
mean[:, 0] + 1.96 * np.sqrt(var[:, 0]),
color="C0",
alpha=0.2,
)
plt.plot(xx, samples[:, :, 0].numpy().T, "C0", linewidth=0.5)
_ = plt.xlim(-0.1, 1.1)
GP regression in higher dimensions#
Very little changes when the input space has more than one dimension. By default, the lengthscales is an isotropic (scalar) parameter. It is generally recommended that you allow to tune a different lengthscale for each dimension (Automatic Relevance Determination, ARD): simply initialize lengthscales with an array of length \(D\) corresponding to the input dimension of X. See Manipulating kernels for further information.
Further reading#
Stochastic Variational Inference for scalability with SVGP for cases where there are a large number of observations.
Ordinal regression if the data is ordinal.
Multi-output models and coregionalisation if
Yis multidimensional.