Manipulating GPflow models#
One of the key ingredients in GPflow is the model class, which enables you to carefully control parameters. This notebook shows how some of these parameter control features work, and how to build your own model with GPflow. First we’ll look at:
how to view models and parameters
how to set parameter values
how to constrain parameters (for example, variance > 0)
how to fix model parameters
how to apply priors to parameters
how to optimize models
Then we’ll show how to build a simple logistic regression model, demonstrating the ease of the parameter framework.
GPy users should feel right at home, but there are some small differences.
First, let’s deal with the usual notebook boilerplate and make a simple GP regression model. See Basic (Gaussian likelihood) GP regression model for specifics of the model; we just want some parameters to play with.
[1]:
import numpy as np
import tensorflow_probability as tfp
import gpflow
from gpflow.utilities import print_summary, set_trainable, to_default_float
2022-10-13 08:59:08.297206: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
2022-10-13 08:59:08.421420: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcudart.so.11.0'; dlerror: libcudart.so.11.0: cannot open shared object file: No such file or directory
2022-10-13 08:59:08.421458: I tensorflow/stream_executor/cuda/cudart_stub.cc:29] Ignore above cudart dlerror if you do not have a GPU set up on your machine.
2022-10-13 08:59:08.450686: E tensorflow/stream_executor/cuda/cuda_blas.cc:2981] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
2022-10-13 08:59:09.023057: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer.so.7'; dlerror: libnvinfer.so.7: cannot open shared object file: No such file or directory
2022-10-13 08:59:09.023159: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libnvinfer_plugin.so.7'; dlerror: libnvinfer_plugin.so.7: cannot open shared object file: No such file or directory
2022-10-13 08:59:09.023169: W tensorflow/compiler/tf2tensorrt/utils/py_utils.cc:38] TF-TRT Warning: Cannot dlopen some TensorRT libraries. If you would like to use Nvidia GPU with TensorRT, please make sure the missing libraries mentioned above are installed properly.
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.decorator.check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.inheritance.inherit_check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
We begin by creating a very simple GP regression model:
[2]:
# generate toy data
np.random.seed(1)
X = np.random.rand(20, 1)
Y = np.sin(12 * X) + 0.66 * np.cos(25 * X) + np.random.randn(20, 1) * 0.01
m = gpflow.models.GPR(
(X, Y), kernel=gpflow.kernels.Matern32() + gpflow.kernels.Linear()
)
/home/circleci/project/gpflow/experimental/utils.py:42: UserWarning: You're calling gpflow.experimental.check_shapes.checker.ShapeChecker.__init__ which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
warn(
2022-10-13 08:59:11.591263: W tensorflow/stream_executor/platform/default/dso_loader.cc:64] Could not load dynamic library 'libcuda.so.1'; dlerror: libcuda.so.1: cannot open shared object file: No such file or directory
2022-10-13 08:59:11.591288: W tensorflow/stream_executor/cuda/cuda_driver.cc:263] failed call to cuInit: UNKNOWN ERROR (303)
2022-10-13 08:59:11.591308: I tensorflow/stream_executor/cuda/cuda_diagnostics.cc:156] kernel driver does not appear to be running on this host (2163c186b2a0): /proc/driver/nvidia/version does not exist
2022-10-13 08:59:11.591547: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.
Viewing, getting, and setting parameters#
You can display the state of the model in a terminal by using print_summary(m). You can change the display format using the fmt keyword argument, e.g. 'html'. In a notebook, you can also use fmt='notebook' or set the default printing format as notebook:
[3]:
print_summary(m, fmt="notebook")
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.kernel.kernels[0].lengthscales | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 1 |
[4]:
gpflow.config.set_default_summary_fmt("notebook")
This model has four parameters. The kernel is made of the sum of two parts. The first (counting from zero) is a Matern32 kernel that has a variance parameter and a lengthscales parameter; the second is a linear kernel that has only a variance parameter. There is also a parameter that controls the variance of the noise, as part of the likelihood.
All the model variables have been initialized at 1.0. You can access individual parameters in the same way that you display the state of the model in a terminal; for example, to see all the parameters that are part of the likelihood, run:
[5]:
print_summary(m.likelihood)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Gaussian.variance | Parameter | Softplus + Shift | True | () | float64 | 1 |
This gets more useful with more complex models!
To set the value of a parameter, just use assign():
[6]:
m.kernel.kernels[0].lengthscales.assign(0.5)
m.likelihood.variance.assign(0.01)
print_summary(m, fmt="notebook")
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.kernel.kernels[0].lengthscales | Parameter | Softplus | True | () | float64 | 0.5 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.01 |
Constraints and trainable variables#
GPflow helpfully creates an unconstrained representation of all the variables. In the previous example, all the variables are constrained positively (see the transform column in the table); the unconstrained representation is given by \(\alpha = \log(\exp(\theta)-1)\). The trainable_parameters property returns the constrained values:
[7]:
m.trainable_parameters
[7]:
(<Parameter: name=softplus, dtype=float64, shape=[], fn="softplus", numpy=0.5>,
<Parameter: name=softplus, dtype=float64, shape=[], fn="softplus", numpy=1.0>,
<Parameter: name=softplus, dtype=float64, shape=[], fn="softplus", numpy=1.0>,
<Parameter: name=chain_of_shift_of_softplus, dtype=float64, shape=[], fn="chain_of_shift_of_softplus", numpy=0.010000000000000002>)
Each parameter has an unconstrained_variable attribute that enables you to access the unconstrained value as a TensorFlow Variable.
[8]:
p = m.kernel.kernels[0].lengthscales
p.unconstrained_variable
[8]:
<tf.Variable 'softplus:0' shape=() dtype=float64, numpy=-0.43275212956718856>
You can also check the unconstrained value as follows:
[9]:
p.transform.inverse(p)
[9]:
<tf.Tensor: shape=(), dtype=float64, numpy=-0.43275212956718856>
Constraints are handled by the Bijector classes from the tensorflow_probability package. You might prefer to use the constraint \(\alpha = \log(\theta)\); this is easily done by replacing the parameter with one that has a different transform attribute (here we make sure to copy all other attributes across from the old parameter; this is not necessary when there is no prior and the trainable state is still the default of True):
[10]:
old_parameter = m.kernel.kernels[0].lengthscales
new_parameter = gpflow.Parameter(
old_parameter,
trainable=old_parameter.trainable,
prior=old_parameter.prior,
name=old_parameter.name.split(":")[
0
], # tensorflow is weird and adds ':0' to the name
transform=tfp.bijectors.Exp(),
)
m.kernel.kernels[0].lengthscales = new_parameter
Though the lengthscale itself remains the same, the unconstrained lengthscale has changed:
[11]:
p.transform.inverse(p)
[11]:
<tf.Tensor: shape=(), dtype=float64, numpy=-0.43275212956718856>
To replace the transform of a parameter you need to recreate the parameter with updated transform:
[12]:
p = m.kernel.kernels[0].variance
m.kernel.kernels[0].variance = gpflow.Parameter(
p.numpy(), transform=tfp.bijectors.Exp()
)
[13]:
print_summary(m, fmt="notebook")
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Exp | True | () | float64 | 1 | |
| GPR.kernel.kernels[0].lengthscales | Parameter | Exp | True | () | float64 | 0.5 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.01 |
Changing whether a parameter will be trained in optimization#
Another helpful feature is the ability to fix parameters. To do this, simply set the trainable attribute to False; this is shown in the trainable column of the representation, and the corresponding variable is removed from the free state.
[14]:
set_trainable(m.kernel.kernels[1].variance, False)
print_summary(m)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Exp | True | () | float64 | 1 | |
| GPR.kernel.kernels[0].lengthscales | Parameter | Exp | True | () | float64 | 0.5 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | False | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.01 |
[15]:
m.trainable_parameters
[15]:
(<Parameter: name=softplus, dtype=float64, shape=[], fn="exp", numpy=0.5>,
<Parameter: name=exp, dtype=float64, shape=[], fn="exp", numpy=1.0>,
<Parameter: name=chain_of_shift_of_softplus, dtype=float64, shape=[], fn="chain_of_shift_of_softplus", numpy=0.010000000000000002>)
To unfix a parameter, just set the trainable attribute to True again.
[16]:
set_trainable(m.kernel.kernels[1].variance, True)
print_summary(m)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Exp | True | () | float64 | 1 | |
| GPR.kernel.kernels[0].lengthscales | Parameter | Exp | True | () | float64 | 0.5 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | True | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.01 |
NOTE: If you want to recursively change the trainable status of an object that contains parameters, you must use the set_trainable() utility function.
A module (e.g. a model, kernel, likelihood, … instance) does not have a trainable attribute:
[17]:
try:
m.kernel.trainable
except AttributeError:
print(f"{m.kernel.__class__.__name__} does not have a trainable attribute")
Sum does not have a trainable attribute
[18]:
set_trainable(m.kernel, False)
print_summary(m)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Exp | False | () | float64 | 1 | |
| GPR.kernel.kernels[0].lengthscales | Parameter | Exp | False | () | float64 | 0.5 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | False | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.01 |
Priors#
You can set priors in the same way as transforms and trainability, by using tensorflow_probability distribution objects. Let’s set a Gamma prior on the variance of the Matern32 kernel.
[19]:
k = gpflow.kernels.Matern32()
k.variance.prior = tfp.distributions.Gamma(
to_default_float(2), to_default_float(3)
)
print_summary(k)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| Matern32.variance | Parameter | Softplus | Gamma | True | () | float64 | 1 |
| Matern32.lengthscales | Parameter | Softplus | True | () | float64 | 1 |
[20]:
m.kernel.kernels[0].variance.prior = tfp.distributions.Gamma(
to_default_float(2), to_default_float(3)
)
print_summary(m)
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| GPR.kernel.kernels[0].variance | Parameter | Exp | Gamma | False | () | float64 | 1 |
| GPR.kernel.kernels[0].lengthscales | Parameter | Exp | False | () | float64 | 0.5 | |
| GPR.kernel.kernels[1].variance | Parameter | Softplus | False | () | float64 | 1 | |
| GPR.likelihood.variance | Parameter | Softplus + Shift | True | () | float64 | 0.01 |
Optimization#
To optimize your model, first create an instance of an optimizer (in this case, gpflow.optimizers.Scipy), which has optional arguments that are passed to scipy.optimize.minimize (we minimize the negative log likelihood). Then, call the minimize method of that optimizer, with your model as the optimization target. Variables that have priors are maximum a priori (MAP) estimated, that is, we add the log prior to the log likelihood, and otherwise use Maximum Likelihood.
[21]:
opt = gpflow.optimizers.Scipy()
opt.minimize(m.training_loss, variables=m.trainable_variables)
[21]:
fun: 27.184339014098025
hess_inv: <1x1 LbfgsInvHessProduct with dtype=float64>
jac: array([-1.02640284e-06])
message: 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
nfev: 8
nit: 7
njev: 8
status: 0
success: True
x: array([-0.36990459])
Building new models#
To build new models, you’ll need to inherit from gpflow.models.BayesianModel. Parameters are instantiated with gpflow.Parameter. You might also be interested in gpflow.Module (a subclass of tf.Module), which acts as a ‘container’ for Parameters (for example, kernels are gpflow.Modules).
In this very simple demo, we’ll implement linear multiclass classification.
There are two parameters: a weight matrix and a bias (offset). You can use Parameter objects directly, like any TensorFlow tensor.
The training objective depends on the type of model; it may be possible to implement the exact (log)marginal likelihood, or only a lower bound to the log marginal likelihood (ELBO). You need to implement this as the maximum_log_likelihood_objective method. The BayesianModel parent class provides a log_posterior_density method that returns the maximum_log_likelihood_objective plus the sum of the log-density of any priors on hyperparameters, which can be used for MCMC. GPflow
provides mixin classes that define a training_loss method that returns the negative of (maximum likelihood objective + log prior density) for MLE/MAP estimation to be passed to optimizer’s minimize method. Models that derive from InternalDataTrainingLossMixin are expected to store the data internally, and their training_loss does not take any arguments and can be passed directly to minimize. Models that take data as an argument to their maximum_log_likelihood_objective
method derive from ExternalDataTrainingLossMixin, which provides a training_loss_closure to take the data and return the appropriate closure for optimizer.minimize. This is also discussed in the GPflow with TensorFlow 2 notebook.
[22]:
import tensorflow as tf
class LinearMulticlass(
gpflow.models.BayesianModel, gpflow.models.InternalDataTrainingLossMixin
):
# The InternalDataTrainingLossMixin provides the training_loss method.
# (There is also an ExternalDataTrainingLossMixin for models that do not encapsulate data.)
def __init__(self, X, Y, name=None):
super().__init__(name=name) # always call the parent constructor
self.X = X.copy() # X is a NumPy array of inputs
self.Y = (
Y.copy()
) # Y is a 1-of-k (one-hot) representation of the labels
self.num_data, self.input_dim = X.shape
_, self.num_classes = Y.shape
# make some parameters
self.W = gpflow.Parameter(
np.random.randn(self.input_dim, self.num_classes)
)
self.b = gpflow.Parameter(np.random.randn(self.num_classes))
# ^^ You must make the parameters attributes of the class for
# them to be picked up by the model. i.e. this won't work:
#
# W = gpflow.Parameter(... <-- must be self.W
def maximum_log_likelihood_objective(self):
p = tf.nn.softmax(
tf.matmul(self.X, self.W) + self.b
) # Parameters can be used like a tf.Tensor
return tf.reduce_sum(
tf.math.log(p) * self.Y
) # be sure to return a scalar
…and that’s it. Let’s build a really simple demo to show that it works.
[23]:
np.random.seed(123)
X = np.vstack(
[
np.random.randn(10, 2) + [2, 2],
np.random.randn(10, 2) + [-2, 2],
np.random.randn(10, 2) + [2, -2],
]
)
Y = np.repeat(np.eye(3), 10, 0)
import matplotlib.pyplot as plt
plt.style.use("ggplot")
%matplotlib inline
plt.rcParams["figure.figsize"] = (12, 6)
_ = plt.scatter(
X[:, 0], X[:, 1], 100, np.argmax(Y, 1), lw=2, cmap=plt.cm.viridis
)
[24]:
m = LinearMulticlass(X, Y)
m
[24]:
| name | class | transform | prior | trainable | shape | dtype | value |
|---|---|---|---|---|---|---|---|
| LinearMulticlass.W | Parameter | Identity | True | (2, 3) | float64 | [[-0.77271, 0.79486, 0.31427... | |
| LinearMulticlass.b | Parameter | Identity | True | (3,) | float64 | [ 0.04549 -0.23309 -1.1983 ] |
[25]:
opt = gpflow.optimizers.Scipy()
opt.minimize(m.training_loss, variables=m.trainable_variables)
[25]:
fun: 1.2560984620758726e-05
hess_inv: <9x9 LbfgsInvHessProduct with dtype=float64>
jac: array([ 4.28392990e-06, 1.15665823e-06, -5.44058813e-06, -2.77570188e-06,
2.97110223e-06, -1.95400347e-07, -8.81900274e-07, 2.52856127e-06,
-1.64666099e-06])
message: 'CONVERGENCE: NORM_OF_PROJECTED_GRADIENT_<=_PGTOL'
nfev: 27
nit: 26
njev: 27
status: 0
success: True
x: array([ 8.55849743, -30.63655328, 22.4144818 , 23.79332963,
21.27896803, -44.17402754, 11.85784428, -12.94743432,
-0.29631308])
[26]:
xx, yy = np.mgrid[-4:4:200j, -4:4:200j]
X_test = np.vstack([xx.flatten(), yy.flatten()]).T
f_test = np.dot(X_test, m.W.numpy()) + m.b.numpy()
p_test = np.exp(f_test)
p_test /= p_test.sum(1)[:, None]
[27]:
plt.figure(figsize=(12, 6))
for i in range(3):
plt.contour(
xx, yy, p_test[:, i].reshape(200, 200), [0.5], colors="k", linewidths=1
)
_ = plt.scatter(
X[:, 0], X[:, 1], 100, np.argmax(Y, 1), lw=2, cmap=plt.cm.viridis
)
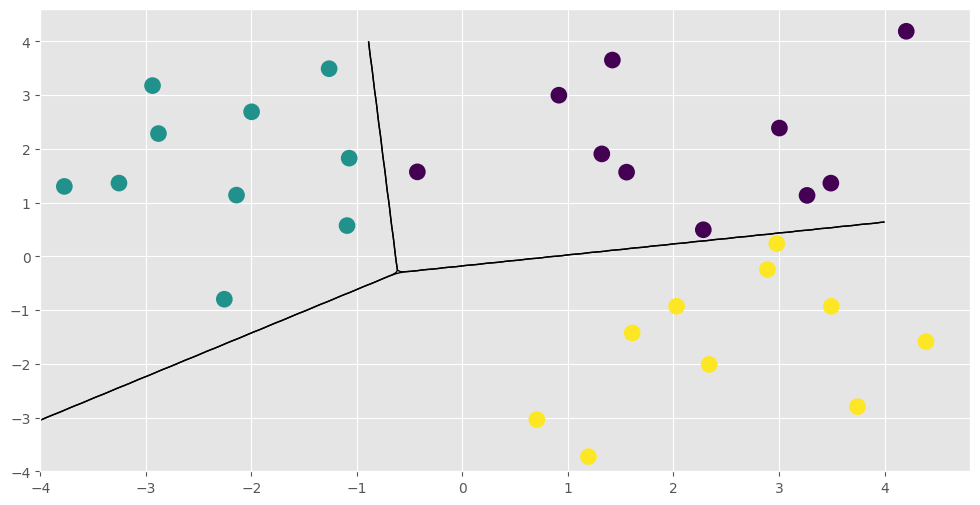
That concludes the new model example and this notebook. You might want to see for yourself that the LinearMulticlass model and its parameters have all the functionality demonstrated here. You could also add some priors and run Hamiltonian Monte Carlo using the HMC optimizer gpflow.train.HMC and its sample method. See Markov Chain Monte Carlo (MCMC) for more information on running the sampler.